A GATA factor mediates cell type-restricted induction of HLA-E gene transcription by gamma interferon
- PMID: 15226423
- PMCID: PMC434230
- DOI: 10.1128/MCB.24.14.6194-6204.2004
A GATA factor mediates cell type-restricted induction of HLA-E gene transcription by gamma interferon
Abstract
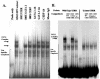
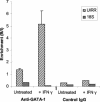
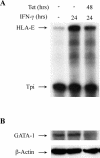
The human major histocompatibility complex (MHC) class Ib gene, HLA-E, codes for the major ligand of the inhibitory receptor NK-G-2A, which is present on most natural killer (NK) cells and some CD8(+) cytotoxic T lymphocytes. We have previously shown that gamma interferon (IFN-gamma) induction of HLA-E gene transcription is mediated through a distinct IFN-gamma-responsive element, the IFN response region (IRR), in all cell types studied. We have now identified and characterized a cell type-restricted enhancer of IFN-gamma-mediated induction of HLA-E gene transcription, designated the upstream interferon response region (UIRR), which is located immediately upstream of the IRR. The UIRR mediates a three- to eightfold enhancement of IFN-gamma induction of HLA-E transcription in some cell lines but not in others, and it functions only in the presence of an adjacent IRR. The UIRR contains a variant GATA binding site (AGATAC) that is critical to both IFN-gamma responsiveness and to the formation of a specific binding complex containing GATA-1 in K562 cell nuclear extracts. The binding of GATA-1 to this site in response to IFN-gamma was confirmed in vivo in a chromatin immunoprecipitation assay. Forced expression of GATA-1 in nonexpressing U937 cells resulted in a four- to fivefold enhancement of the IFN-gamma response from HLA-E promoter constructs containing a wild-type but not a GATA-1 mutant UIRR sequence and increased the IFN-gamma response of the endogenous HLA-E gene. Knockdown of GATA-1 expression in K562 cells resulted in a approximately 4-fold decrease in the IFN-gamma response of the endogenous HLA-E gene, consistent with loss of the increase in IFN-gamma response of HLA-E promoter-driven constructs containing the UIRR in wild-type K562 cells. Coexpression of wild-type and mutant adenovirus E1a proteins that sequester p300/CBP eliminated IFN-gamma-mediated enhancement through the UIRR, but only partially reduced induction through the IRR, implicating p300/CBP binding to Stat-1alpha at the IRR in the recruitment of GATA-1 to mediate the cooperation between the UIRR and IRR. We propose that the GATA-1 transcription factor represents a cell type-restricted mediator of IFN-gamma induction of the HLA-E gene.
Figures





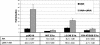

References
-
- Adams, E. J., and P. Parham. 2001. Species-specific evolution of MHC class I genes in the higher primates. Immunol. Rev. 183:41-64. - PubMed
-
- Aittomaki, S., J. Yang, E. W. Scott, M. C. Simon, and O. Silvennoimen. 2002. Distinct functions for signal transducer and activator of transcription 1 and PU. 1 in transcriptional activation of Fcγ receptor 1 promoter. Blood 100:1078-1080. - PubMed
-
- Aldrich, C. J., A. DeCloux, A. S. Woods, R. J. Cotter, M. J. Soloski, and J. Forman. 1994. Identification of a Tap-dependent leader peptide recognized by alloreactive T cells specific for a class Ib antigen. Cell 79:649-658. - PubMed
-
- Bach, E. A., M. Aguet, and R. D. Schrieber. 1997. The IFN-gamma receptor: a paradigm for cytokine receptor signaling. Annu. Rev. Immunol. 15:563-591. - PubMed
-
- Barrett, D. 2003. Ph.D. thesis. Virginia Commonwealth University, Richmond.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
