Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts
- PMID: 15226427
- PMCID: PMC434237
- DOI: 10.1128/MCB.24.14.6241-6252.2004
Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts
Abstract
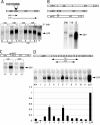
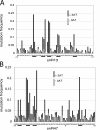
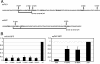
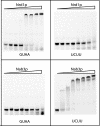
RNA polymerase II (Pol II) termination is triggered by sequences present in the nascent transcript. Termination of pre-mRNA transcription is coupled to recognition of cis-acting sequences that direct cleavage and polyadenylation of the pre-mRNA. Termination of nonpolyadenylated [non-poly(A)] Pol II transcripts in Saccharomyces cerevisiae requires the RNA-binding proteins Nrd1 and Nab3. We have used a mutational strategy to characterize non-poly(A) termination elements downstream of the SNR13 and SNR47 snoRNA genes. This approach detected two common RNA sequence motifs, GUA[AG] and UCUU. The first motif corresponds to the known Nrd1-binding site, which we have verified here by gel mobility shift assays. We also show that Nab3 protein binds specifically to RNA containing the UCUU motif. Taken together, our data suggest that Nrd1 and Nab3 binding sites play a significant role in defining non-poly(A) terminators. As is the case with poly(A) terminators, there is no strong consensus for non-poly(A) terminators, and the arrangement of Nrd1p and Nab3p binding sites varies considerably. In addition, the organization of these sequences is not strongly conserved among even closely related yeasts. This indicates a large degree of genetic variability. Despite this variability, we were able to use a computational model to show that the binding sites for Nrd1 and Nab3 can identify genes for which transcription termination is mediated by these proteins.
Figures




References
-
- Antson, A. A. 2000. Single-stranded-RNA binding proteins. Curr. Opin. Struct. Biol. 10:87-94. - PubMed
-
- Bentley, D. 2002. The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14:336-342. - PubMed
-
- Birse, C. E., L. Minvielle-Sebastia, B. A. Lee, W. Keller, and N. J. Proudfoot. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280:298-301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
