The E4F protein is required for mitotic progression during embryonic cell cycles
- PMID: 15226446
- PMCID: PMC434264
- DOI: 10.1128/MCB.24.14.6467-6475.2004
The E4F protein is required for mitotic progression during embryonic cell cycles
Abstract
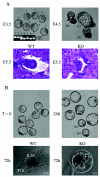
The ubiquitously expressed E4F protein was originally identified as an E1A-regulated cellular transcription factor required for adenovirus replication. The function of this protein in normal cell physiology remains largely unknown. To address this issue, we generated E4F knockout mice by gene targeting. Embryos lacking E4F die at the peri-implantation stage, while in vitro-cultured E4F(-/-) blastocysts exhibit defects in mitotic progression, chromosomal missegregation, and increased apoptosis. Consistent with these observations, we found that E4F localizes to the mitotic spindle during the M phase of early embryos. Our results establish a crucial role for E4F during early embryonic cell cycles and reveal an unexpected function for E4F in mitosis.
Figures





References
-
- Cleveland, D. W., Y. Mao, and K. F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407-421. - PubMed
-
- Colombo, R., G. F. Draetta, and S. Chiocca. 2003. Modulation of p120E4F transcriptional activity by the Gam1 adenoviral early protein. Oncogene 22:2541-2547. - PubMed
-
- Cutts, S. M., K. J. Fowler, B. T. Kile, L. L. Hii, R. A. O'Dowd, D. F. Hudson, R. Saffery, P. Kalitsis, E. Earle, and K. H. Choo. 1999. Defective chromosome segregation, microtubule bundling and nuclear bridging in inner centromere protein gene (Incenp)-disrupted mice. Hum. Mol. Genet. 8:1145-1155. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
