Characterization of the polyene macrolide P450 epoxidase from Streptomyces natalensis that converts de-epoxypimaricin into pimaricin
- PMID: 15228385
- PMCID: PMC1134766
- DOI: 10.1042/BJ20040490
Characterization of the polyene macrolide P450 epoxidase from Streptomyces natalensis that converts de-epoxypimaricin into pimaricin
Abstract
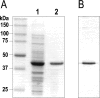
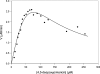
The biosynthesis of the antifungal agent pimaricin by Streptomyces natalensis has been proposed to involve a cytochrome P450 encoded by the gene pimD. Pimaricin is derived from its immediate precursor de-epoxypimaricin by epoxidation of the C-4-C-5 double bond on the macrolactone ring. We have overproduced PimD with a N-terminal His6 affinity tag in Escherichia coli and purified the enzyme for kinetic analysis. The protein showed a reduced CO-difference spectrum with a Soret maximum at 450 nm, indicating that it is a cytochrome P450. Purified PimD was shown to catalyse the in vitro C-4-C-5 epoxidation of 4,5-de-epoxypimaricin to pimaricin. The enzyme was dependent on NADPH for activity with optimal pH at 7.5, and the temperature optimum was 30 degrees C. The kcat value for the epoxidation of de-epoxypimaricin was similar to the values reported for other macrolide oxidases. Enzyme activity was inhibited at high substrate concentration. This is the first time that a polyene macrolide P450 mono-oxygenase has been expressed heterologously and studied. The unique specificity of this epoxidase should be useful for the oxidative modification of novel polyene macrolide antibiotics.
Figures


Similar articles
-
Engineered biosynthesis of novel polyenes: a pimaricin derivative produced by targeted gene disruption in Streptomyces natalensis.Chem Biol. 2001 Jul;8(7):635-44. doi: 10.1016/s1074-5521(01)00033-3. Chem Biol. 2001. PMID: 11451665
-
Characterization of the macrolide P-450 hydroxylase from Streptomyces venezuelae which converts narbomycin to picromycin.Biochemistry. 1998 Oct 20;37(42):14937-42. doi: 10.1021/bi981699c. Biochemistry. 1998. PMID: 9778370
-
Isolation and characterization of pcsB, the gene for a polyene carboxamide synthase that tailors pimaricin into AB-400.Appl Microbiol Biotechnol. 2010 Feb;85(6):1809-19. doi: 10.1007/s00253-009-2195-1. Epub 2009 Aug 26. Appl Microbiol Biotechnol. 2010. PMID: 19707754
-
Biosynthesis of the polyene macrolide antibiotic nystatin in Streptomyces noursei.Appl Microbiol Biotechnol. 2005 Jun;67(4):436-43. doi: 10.1007/s00253-004-1802-4. Epub 2005 Feb 8. Appl Microbiol Biotechnol. 2005. PMID: 15700127 Review.
-
Biotechnological production and application of the antibiotic pimaricin: biosynthesis and its regulation.Appl Microbiol Biotechnol. 2016 Jan;100(1):61-78. doi: 10.1007/s00253-015-7077-0. Epub 2015 Oct 29. Appl Microbiol Biotechnol. 2016. PMID: 26512010 Free PMC article. Review.
Cited by
-
Cooperative Substrate Binding Controls Catalysis in Bacterial Cytochrome P450terp (CYP108A1).J Am Chem Soc. 2023 Feb 13:10.1021/jacs.2c12388. doi: 10.1021/jacs.2c12388. Online ahead of print. J Am Chem Soc. 2023. PMID: 36779970 Free PMC article.
-
Genome Sequence-Guided Finding of Lucensomycin Production by Streptomyces achromogenes Subsp. streptozoticus NBRC14001.Microorganisms. 2021 Dec 26;10(1):37. doi: 10.3390/microorganisms10010037. Microorganisms. 2021. PMID: 35056487 Free PMC article.
-
An Unprecedented Number of Cytochrome P450s Are Involved in Secondary Metabolism in Salinispora Species.Microorganisms. 2022 Apr 21;10(5):871. doi: 10.3390/microorganisms10050871. Microorganisms. 2022. PMID: 35630316 Free PMC article.
-
Functional analysis of MycCI and MycG, cytochrome P450 enzymes involved in biosynthesis of mycinamicin macrolide antibiotics.Chem Biol. 2008 Sep 22;15(9):950-9. doi: 10.1016/j.chembiol.2008.07.014. Chem Biol. 2008. PMID: 18804032 Free PMC article.
-
Proximal ligand electron donation and reactivity of the cytochrome P450 ferric-peroxo anion.J Am Chem Soc. 2012 Apr 18;134(15):6673-84. doi: 10.1021/ja211499q. Epub 2012 Apr 4. J Am Chem Soc. 2012. PMID: 22444582 Free PMC article.
References
-
- Munro A. W., Lindsay J. G. Bacterial cytochromes P-450. Mol. Microbiol. 1996;20:1115–1125. - PubMed
-
- Haydock S. F., Dowson J. A., Dhillon N., Roberts G. A., Cortés J., Leadlay P. F. Cloning and sequencing analysis of genes involved in erythromycin biosynthesis in Saccharopolyspora erythraea: sequence similarities between EryG and a family of S-adenosylmethionine-dependent methyltransferases. Mol. Gen. Genet. 1991;230:120–128. - PubMed
-
- Merson-Davies L. A., Cundliffe E. Analysis of five tylosin biosynthetic genes from the tylIBA region of Streptomyces fradiae genome. Mol. Microbiol. 1994;13:349–355. - PubMed
-
- Rodríguez A. M., Olano C., Méndez C., Hutchinson C. R., Salas J. A. A cytochrome P450-like gene possibly involved in oleandomycin biosynthesis by Streptomyces antibioticus. FEMS Microbiol. Lett. 1995;127:117–120. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

