Substrate specificities of bacterial and human AlkB proteins
- PMID: 15229293
- PMCID: PMC443531
- DOI: 10.1093/nar/gkh655
Substrate specificities of bacterial and human AlkB proteins
Abstract
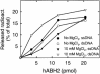
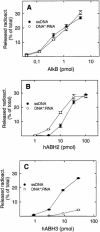
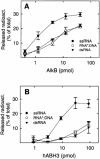
Methylating agents introduce cytotoxic 1-methyladenine (1-meA) and 3-methylcytosine (3-meC) residues into nucleic acids, and it was recently demonstrated that the Escherichia coli AlkB protein and two human homologues, hABH2 and hABH3, can remove these lesions from DNA by oxidative demethylation. Moreover, AlkB and hABH3 were also found to remove 1-meA and 3-meC from RNA, suggesting that cellular RNA repair can occur. We have here studied the preference of AlkB, hABH2 and hABH3 for single-stranded DNA (ssDNA) or double-stranded DNA (dsDNA), and show that AlkB and hABH3 prefer ssDNA, while hABH2 prefers dsDNA. This was consistently observed with three different oligonucleotide substrates, implying that the specificity for single-stranded versus double-stranded DNA is sequence independent. The dsDNA preference of hABH2 was observed only in the presence of magnesium. The activity of the enzymes on single-stranded RNA (ssRNA), double-stranded RNA (dsRNA) and DNA/RNA hybrids was also investigated, and the results generally confirm the notion that while AlkB and hABH3 tend to prefer single-stranded nucleic acids, hABH2 is more active on double-stranded substrates. These results may contribute to identifying the main substrates of bacterial and human AlkB proteins in vivo.
Figures

Similar articles
-
AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation.Mol Cell. 2004 Oct 8;16(1):107-16. doi: 10.1016/j.molcel.2004.09.002. Mol Cell. 2004. PMID: 15469826
-
Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA.Nature. 2003 Feb 20;421(6925):859-63. doi: 10.1038/nature01363. Nature. 2003. PMID: 12594517
-
Interaction of human and bacterial AlkB proteins with DNA as probed through chemical cross-linking studies.Nucleic Acids Res. 2004 Mar 5;32(4):1548-54. doi: 10.1093/nar/gkh319. Print 2004. Nucleic Acids Res. 2004. PMID: 15004242 Free PMC article.
-
AlkB demethylases flip out in different ways.DNA Repair (Amst). 2008 Nov 1;7(11):1916-23. doi: 10.1016/j.dnarep.2008.07.015. Epub 2008 Sep 6. DNA Repair (Amst). 2008. PMID: 18723127 Review.
-
Alkylation damage in DNA and RNA--repair mechanisms and medical significance.DNA Repair (Amst). 2004 Nov 2;3(11):1389-407. doi: 10.1016/j.dnarep.2004.05.004. DNA Repair (Amst). 2004. PMID: 15380096 Review.
Cited by
-
DNA Demethylation in the Processes of Repair and Epigenetic Regulation Performed by 2-Ketoglutarate-Dependent DNA Dioxygenases.Int J Mol Sci. 2021 Sep 29;22(19):10540. doi: 10.3390/ijms221910540. Int J Mol Sci. 2021. PMID: 34638881 Free PMC article. Review.
-
Dysregulation of RNA modification systems in clinical populations with neurocognitive disorders.Neural Regen Res. 2024 Jun 1;19(6):1256-1261. doi: 10.4103/1673-5374.385858. Epub 2023 Sep 22. Neural Regen Res. 2024. PMID: 37905873 Free PMC article.
-
Spectroscopic and in vitro Investigations of Fe2+ /α-Ketoglutarate-Dependent Enzymes Involved in Nucleic Acid Repair and Modification.Chembiochem. 2022 Jun 3;23(11):e202100605. doi: 10.1002/cbic.202100605. Epub 2022 Feb 15. Chembiochem. 2022. PMID: 35040547 Free PMC article. Review.
-
The DNA dioxygenase ALKBH2 protects Arabidopsis thaliana against methylation damage.Nucleic Acids Res. 2012 Aug;40(14):6620-31. doi: 10.1093/nar/gks327. Epub 2012 Apr 24. Nucleic Acids Res. 2012. PMID: 22532610 Free PMC article.
-
Pediatric brain tumors: mutations of two dioxygenases (hABH2 and hABH3) that directly repair alkylation damage.J Neurooncol. 2009 Sep;94(2):195-201. doi: 10.1007/s11060-009-9837-0. Epub 2009 Mar 17. J Neurooncol. 2009. PMID: 19290481
References
-
- Sedgwick B. and Lindahl,T. (2002) Recent progress on the Ada response for inducible repair of DNA alkylation damage. Oncogene, 21, 8886–8894. - PubMed
-
- Begley T.J. and Samson,L.D. (2003) AlkB mystery solved: oxidative demethylation of N1-methyladenine and N3-methylcytosine adducts by a direct reversal mechanism. Trends Biochem. Sci., 28, 2–5. - PubMed
-
- Schofield C.J. and Zhang,Z. (1999) Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Curr. Opin. Struct. Biol., 9, 722–731. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

