mlstdbNet - distributed multi-locus sequence typing (MLST) databases
- PMID: 15230973
- PMCID: PMC459212
- DOI: 10.1186/1471-2105-5-86
mlstdbNet - distributed multi-locus sequence typing (MLST) databases
Abstract
Background: Multi-locus sequence typing (MLST) is a method of typing that facilitates the discrimination of microbial isolates by comparing the sequences of housekeeping gene fragments. The mlstdbNet software enables the implementation of distributed web-accessible MLST databases that can be linked widely over the Internet.
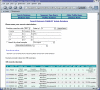
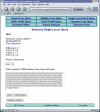
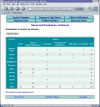
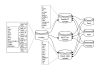
Results: The software enables multiple isolate databases to query a single profiles database that contains allelic profile and sequence definitions. This separation enables isolate databases to be established by individual laboratories, each customized to the needs of the particular project and with appropriate access restrictions, while maintaining the benefits of a single definitive source of profile and sequence information. Databases are described by an XML file that is parsed by a Perl CGI script. The software offers a large number of ways to query the databases and to further break down and export the results generated. Additional features can be enabled by installing third-party (freely available) tools.
Conclusion: Development of a distributed structure for MLST databases offers scalability and flexibility, allowing participating centres to maintain ownership of their own data, without introducing duplication and data integrity issues.
Figures
References
-
- Maiden MCJ, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. - DOI - PMC - PubMed
-
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, Lehvaslaiho H, Matsalla C, Mungall CJ, Osborne BI, Pocock MR, Schattner P, Senger M, Stein LD, Stupka E, Wilkinson MD, Birney E. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

