Chlamydial development is blocked in host cells transfected with Chlamydophila caviae incA
- PMID: 15230981
- PMCID: PMC459217
- DOI: 10.1186/1471-2180-4-24
Chlamydial development is blocked in host cells transfected with Chlamydophila caviae incA
Abstract
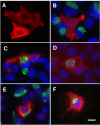
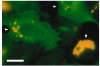
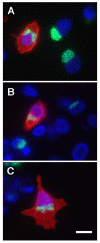
Background: Chlamydiae produce a set of proteins, termed Inc proteins, that are localized to the inclusion membrane and exposed to the host cell cytosol. Little information exists regarding the interaction of Inc proteins with the eukaryotic cell. To examine these interactions, Vaccinia virus vectors and mammalian plasmid-based systems were used to express inc genes in mammalian cells.
Results: Cells transfected with plasmids expressing Chlamydophila caviae incA were not productively infected by C. caviae. Expression of C. caviae incA also reduced inclusion formation by Chlamydia trachomatis, but not to the degree seen for C. caviae. Chlamydia trachomatis incA did not block development of either C. trachomatis or C. caviae. Deletion mutagenesis was used to demonstrate that plasmids encoding either the amino or carboxy-terminal regions of the protein, as well as the changing of a single amino acid within IncA (serine 17) could not block C. caviae infection. Immunoblot analysis of truncated IncA in a Vaccinia virus system provided evidence that serine 17 of C. caviae IncA is a target for phosphorylation.
Conclusions: These experiments provide insight into the interaction of Inc proteins with the host cell and introduce a model system where these interactions can be explored further.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

