U2AF homology motifs: protein recognition in the RRM world
- PMID: 15231733
- PMCID: PMC2043112
- DOI: 10.1101/gad.1206204
U2AF homology motifs: protein recognition in the RRM world
Abstract
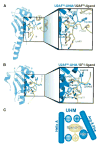
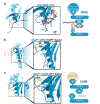
Recent structures of the heterodimeric splicing factor U2 snRNP auxiliary factor (U2AF) have revealed two unexpected examples of RNA recognition motif (RRM)-like domains with specialized features for protein recognition. These unusual RRMs, called U2AF homology motifs (UHMs), represent a novel class of protein recognition motifs. Defining a set of rules to distinguish traditional RRMs from UHMs is key to identifying novel UHM family members. Here we review the critical sequence features necessary to mediate protein-UHM interactions, and perform comprehensive database searches to identify new members of the UHM family. The resulting implications for the functional and evolutionary relationships among candidate UHM family members are discussed.
Figures



References
-
- Abovich N, Liao XC, Rosbash M. The yeast MUD2 protein: An interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes & Dev. 1994;8:843–854. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
