Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27
- PMID: 15231737
- PMCID: PMC443521
- DOI: 10.1101/gad.1200204
Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27
Abstract
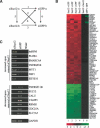
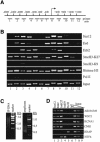
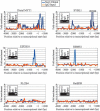
Polycomb group (PcG) complexes 2 and 3 are involved in transcriptional silencing. These complexes contain a histone lysine methyltransferase (HKMT) activity that targets different lysine residues on histones H1 or H3 in vitro. However, it is not known if these histones are methylation targets in vivo because the human PRC2/3 complexes have not been studied in the context of a natural promoter because of the lack of known target genes. Here we report the use of RNA expression arrays and CpG-island DNA arrays to identify and characterize human PRC2/3 target genes. Using oligonucleotide arrays, we first identified a cohort of genes whose expression changes upon siRNA-mediated removal of Suz12, a core component of PRC2/3, from colon cancer cells. To determine which of the putative target genes are directly bound by Suz12 and to precisely map the binding of Suz12 to those promoters, we combined a high-resolution chromatin immunoprecipitation (ChIP) analysis with custom oligonucleotide promoter arrays. We next identified additional putative Suz12 target genes by using ChIP coupled to CpG-island microarrays. We showed that HKMT-Ezh2 and Eed, two other components of the PRC2/3 complexes, colocalize to the target promoters with Suz12. Importantly, recruitment of Suz12, Ezh2 and Eed to target promoters coincides with methylation of histone H3 on Lys 27.
Figures





References
-
- Barnett M.W., Seville, R.A., Nijjar, S., Old, R.W., and Jones, E.A. 2001. Xenopus Enhancer of zeste (XEZ); an anteriorly restricted polycomb gene with a role in neural patterning. Mech. Dev. 102: 157-167. - PubMed
-
- Bellefroid E.J., Bourguignon, C., Hollemann, T., Ma, Q., Anderson, D.J., Kintner, C., and Pieler, T. 1996. X-MyT1, a Xenopus C2HC-type zinc finger protein with a regulatory function in neuronal differentiation. Cell 87: 1191-1202. - PubMed
-
- Bienz M. and Clevers, H. 2000. Linking colorectal cancer to Wnt signaling. Cell 103: 311-320. - PubMed
-
- Bloyer S., Cavalli, G., Brock, H.W., and Dura, J.M. 2003. Identification and characterization of polyhomeotic PREs and TREs. Dev. Biol. 261: 426-442. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
