Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport
- PMID: 15231740
- PMCID: PMC443524
- DOI: 10.1101/gad.1194004
Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport
Abstract
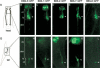
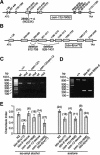
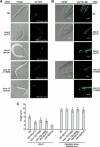
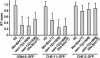
Bardet-Biedl syndrome (BBS) is a genetically heterogeneous developmental disorder whose molecular basis is largely unknown. Here, we show that mutations in the Caenorhabditis elegans bbs-7 and bbs-8 genes cause structural and functional defects in cilia. C. elegans BBS proteins localize predominantly at the base of cilia, and like proteins involved in intraflagellar transport (IFT), a process necessary for cilia biogenesis and maintenance, move bidirectionally along the ciliary axoneme. Importantly, we demonstrate that BBS-7 and BBS-8 are required for the normal localization/motility of the IFT proteins OSM-5/Polaris and CHE-11, and to a notably lesser extent, CHE-2. We propose that BBS proteins play important, selective roles in the assembly and/or function of IFT particle components. Our findings also suggest that some of the cardinal and secondary symptoms of BBS, such as obesity, diabetes, cardiomyopathy, and learning defects may result from cilia dysfunction.
Figures

References
-
- Ansley S.J., Badano, J.L., Blacque, O.E., Hill, J., Hoskins, B.E., Leitch, C.C., Kim, J.C., Ross, A.J., Eichers, E.R., Teslovich, T.M., et al. 2003. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature 425: 628-633. - PubMed
-
- Bale A.E. 2002. Hedgehog signaling and human disease. Annu. Rev. Genomics Hum. Genet. 3: 47-65. - PubMed
-
- Bargmann C.I., Hartwieg, E., and Horvitz, H.R. 1993. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74: 515-527. - PubMed
-
- Blatch G.L. and Lassle, M. 1999. The tetratricopeptide repeat: A structural motif mediating protein–protein interactions. Bioessays 21: 932-939. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
