A protein interaction framework for human mRNA degradation
- PMID: 15231747
- PMCID: PMC442147
- DOI: 10.1101/gr.2122004
A protein interaction framework for human mRNA degradation
Abstract
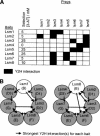
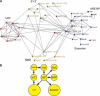
The degradation of mRNA is an important regulatory step in the control of gene expression. However, mammalian RNA decay pathways remain poorly characterized. To provide a framework for studying mammalian RNA decay, a two-hybrid protein interaction map was generated using 54 constructs from 38 human proteins predicted to function in mRNA decay. The results provide evidence for interactions between many different proteins required for mRNA decay. Of particular interest are interactions between the poly(A) ribonuclease and the exosome and between the Lsm complex, decapping factors, and 5'-->3' exonucleases. Moreover, multiple interactions connect 5'-->3' and 3'-->5' decay proteins to each other and to nonsense-mediated decay factors, providing the opportunity for coordination between decay pathways. The interaction network also predicts the internal organization of the exosome and Lsm complexes. Additional interactions connect mRNA decay factors to many novel proteins and to proteins required for other steps in gene expression. These results provide an experimental insight into the organization of proteins required for mRNA decay and their coupling to other cellular processes, and the physiological relevance of many of these interactions are supported by their evolutionary conservation. The interactions also provide a wealth of hypotheses to guide future research on mRNA degradation and demonstrate the power of exhaustive protein interaction mapping in aiding understanding of uncharacterized protein complexes and pathways.
Copyright 2004 Cold Spring Harbor Laboratory Press ISSN
Figures


References
-
- Chen, C.Y., Gherzi, R., Ong, S.E., Chan, E.L., Raijmakers, R., Pruijn, G.J., Stoecklin, G., Moroni, C., Mann, M., and Karin, M. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464. - PubMed
-
- Collins, B.M., Cubeddu, L., Naidoo, N., Harrop, S.J., Kornfeld, G.D., Dawes, I.W., Curmi, P.M., and Mabbutt, B.C. 2003. Homomeric ring assemblies of eukaryotic Sm proteins have affinity for both RNA and DNA. Crystal structure of an oligomeric complex of yeast SmF. J. Biol. Chem. 278: 17291–17298. - PubMed
WEB SITE REFERENCES
-
- http://maine.ebi.ac.uk:8000/services/biolayout; Biolayout software.
-
- http://www.hgmp.mrc.ac.uk/Research/RNA; descriptions of all interacting proteins isolated in this study.
-
- http://inparanoid.cgb.ki.se/; The Inparanoid database of pairwise orthologs.
-
- http://www.blueprint.org/bind/bind.php; BIND protein interaction database.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
