Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development
- PMID: 15231776
- PMCID: PMC438629
- DOI: 10.1128/JB.186.14.4449-4456.2004
Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development
Abstract
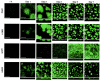
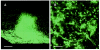
An analysis of the Pseudomonas aeruginosa genomic sequence revealed three gene clusters, PA1381-1393, PA2231-2240, and PA3552-3558, in addition to the alginate biosynthesis gene cluster, which appeared to encode functions for exopolysaccharide (EPS) biosynthesis. Recent evidence indicates that alginate is not a significant component of the extracellular matrix in biofilms of the sequenced P. aeruginosa strain PAO1. We hypothesized that at least one of the three potential EPS gene clusters revealed by genomic sequencing is an important component of P. aeruginosa PAO1 biofilms. Thus, we constructed mutants with chromosomal insertions in PA1383, PA2231, and PA3552. The mutant with a PA2231 defect formed thin unstructured abnormal biofilms. The PA3552 mutant formed structured biofilms that appeared different from those formed by the parent, and the PA1383 mutant formed structured biofilms that were indistinguishable from those formed by the parent. Consistent with a previous report, we found that polysaccharides were one component of the extracellular matrix, which also contained DNA. We suggest that the genes that were inactivated in our PA2231 mutant are required for the production of an EPS, which, although it may be a minor constituent of the matrix, is critical for the formation of P. aeruginosa PAO1 biofilms.
Copyright 2004 American Society for Microbiology
Figures
References
-
- Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1997. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, Inc., New York, N.Y.
-
- Becher, A., and H. P. Schweizer. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. BioTechniques 29:948-950, 952. - PubMed
-
- Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. - PubMed
-
- Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

