Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix
- PMID: 15231777
- PMCID: PMC438632
- DOI: 10.1128/JB.186.14.4457-4465.2004
Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix
Abstract
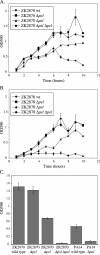
Pseudomonas aeruginosa forms biofilms, which are cellular aggregates encased in an extracellular matrix. Molecular genetics studies of three common autoaggregative phenotypes, namely wrinkled colonies, pellicles, and solid-surface-associated biofilms, led to the identification of two loci, pel and psl, that are involved in the production of carbohydrate-rich components of the biofilm matrix. The pel gene cluster is involved in the production of a glucose-rich matrix material in P. aeruginosa strain PA14 (L. Friedman and R. Kolter, Mol. Microbiol. 51:675-690, 2004). Here we investigate the role of the pel gene cluster in P. aeruginosa strain ZK2870 and identify a second genetic locus, termed psl, involved in the production of a mannose-rich matrix material. The 11 predicted protein products of the psl genes are homologous to proteins involved in carbohydrate processing. P. aeruginosa is thus able to produce two distinct carbohydrate-rich matrix materials. Either carbohydrate-rich matrix component appears to be sufficient for mature biofilm formation, and at least one of them is required for mature biofilm formation in P. aeruginosa strains PA14 and ZK2870.
Copyright 2004 American Society for Microbiology
Figures




Similar articles
-
Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation.J Bacteriol. 2004 Jul;186(14):4466-75. doi: 10.1128/JB.186.14.4466-4475.2004. J Bacteriol. 2004. PMID: 15231778 Free PMC article.
-
Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms.Mol Microbiol. 2004 Feb;51(3):675-90. doi: 10.1046/j.1365-2958.2003.03877.x. Mol Microbiol. 2004. PMID: 14731271
-
The Versatile Pseudomonas aeruginosa Biofilm Matrix Protein CdrA Promotes Aggregation through Different Extracellular Exopolysaccharide Interactions.J Bacteriol. 2020 Sep 8;202(19):e00216-20. doi: 10.1128/JB.00216-20. Print 2020 Sep 8. J Bacteriol. 2020. PMID: 32661078 Free PMC article.
-
Role of polysaccharides in Pseudomonas aeruginosa biofilm development.Curr Opin Microbiol. 2007 Dec;10(6):644-8. doi: 10.1016/j.mib.2007.09.010. Epub 2007 Nov 5. Curr Opin Microbiol. 2007. PMID: 17981495 Free PMC article. Review.
-
[Pseudomonas aeruginosa: characteristics of biofilm process].Mol Gen Mikrobiol Virusol. 2012;(1):3-8. Mol Gen Mikrobiol Virusol. 2012. PMID: 22702137 Review. Russian.
Cited by
-
Carbon Source and Substrate Surface Affect Biofilm Formation by the Plant-Associated Bacterium Pseudomonas donghuensis P482.Int J Mol Sci. 2024 Jul 30;25(15):8351. doi: 10.3390/ijms25158351. Int J Mol Sci. 2024. PMID: 39125921 Free PMC article.
-
Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated.Environ Microbiol. 2012 Aug;14(8):1995-2005. doi: 10.1111/j.1462-2920.2012.02753.x. Epub 2012 Apr 19. Environ Microbiol. 2012. PMID: 22513190 Free PMC article.
-
Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines.mBio. 2013 Jan 29;4(1):e00526-12. doi: 10.1128/mBio.00526-12. mBio. 2013. PMID: 23362320 Free PMC article.
-
Differentiation and distribution of colistin- and sodium dodecyl sulfate-tolerant cells in Pseudomonas aeruginosa biofilms.J Bacteriol. 2007 Jan;189(1):28-37. doi: 10.1128/JB.00720-06. Epub 2006 Oct 13. J Bacteriol. 2007. PMID: 17041046 Free PMC article.
-
The ExpR/Sin quorum-sensing system controls succinoglycan production in Sinorhizobium meliloti.J Bacteriol. 2007 Oct;189(19):7077-88. doi: 10.1128/JB.00906-07. Epub 2007 Jul 20. J Bacteriol. 2007. PMID: 17644606 Free PMC article.
References
-
- Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. - PMC - PubMed
-
- Chou, F. L., H. C. Chou, Y. S. Lin, B. Y. Yang, N. T. Lin, S. F. Weng, and Y. H. Tseng. 1997. The Xanthomonas campestris gumD gene required for synthesis of xanthan gum is involved in normal pigmentation and virulence in causing black rot. Biochem. Biophys. Res. Commun. 233:265-269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

