Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation
- PMID: 15231778
- PMCID: PMC438565
- DOI: 10.1128/JB.186.14.4466-4475.2004
Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation
Abstract
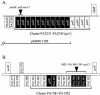
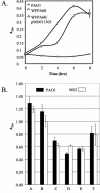
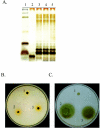
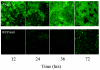
Bacteria inhabiting biofilms usually produce one or more polysaccharides that provide a hydrated scaffolding to stabilize and reinforce the structure of the biofilm, mediate cell-cell and cell-surface interactions, and provide protection from biocides and antimicrobial agents. Historically, alginate has been considered the major exopolysaccharide of the Pseudomonas aeruginosa biofilm matrix, with minimal regard to the different functions polysaccharides execute. Recent chemical and genetic studies have demonstrated that alginate is not involved in the initiation of biofilm formation in P. aeruginosa strains PAO1 and PA14. We hypothesized that there is at least one other polysaccharide gene cluster involved in biofilm development. Two separate clusters of genes with homology to exopolysaccharide biosynthetic functions were identified from the annotated PAO1 genome. Reverse genetics was employed to generate mutations in genes from these clusters. We discovered that one group of genes, designated psl, are important for biofilm initiation. A PAO1 strain with a disruption of the first two genes of the psl cluster (PA2231 and PA2232) was severely compromised in biofilm initiation, as confirmed by static microtiter and continuous culture flow cell and tubing biofilm assays. This impaired biofilm phenotype could be complemented with the wild-type psl sequences and was not due to defects in motility or lipopolysaccharide biosynthesis. These results implicate an as yet unknown exopolysaccharide as being required for the formation of the biofilm matrix. Understanding psl-encoded exopolysaccharide expression and protection in biofilms will provide insight into the pathogenesis of P. aeruginosa in cystic fibrosis and other infections involving biofilms.
Copyright 2004 American Society for Microbiology
Figures
Similar articles
-
The Versatile Pseudomonas aeruginosa Biofilm Matrix Protein CdrA Promotes Aggregation through Different Extracellular Exopolysaccharide Interactions.J Bacteriol. 2020 Sep 8;202(19):e00216-20. doi: 10.1128/JB.00216-20. Print 2020 Sep 8. J Bacteriol. 2020. PMID: 32661078 Free PMC article.
-
Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development.J Bacteriol. 2004 Jul;186(14):4449-56. doi: 10.1128/JB.186.14.4449-4456.2004. J Bacteriol. 2004. PMID: 15231776 Free PMC article.
-
Expression of the psl operon in Pseudomonas aeruginosa PAO1 biofilms: PslA performs an essential function in biofilm formation.Appl Environ Microbiol. 2005 Aug;71(8):4407-13. doi: 10.1128/AEM.71.8.4407-4413.2005. Appl Environ Microbiol. 2005. PMID: 16085831 Free PMC article.
-
Regulation of Biofilm Exopolysaccharide Biosynthesis and Degradation in Pseudomonas aeruginosa.Annu Rev Microbiol. 2022 Sep 8;76:413-433. doi: 10.1146/annurev-micro-041320-111355. Epub 2022 Jun 2. Annu Rev Microbiol. 2022. PMID: 35655342 Review.
-
Role of polysaccharides in Pseudomonas aeruginosa biofilm development.Curr Opin Microbiol. 2007 Dec;10(6):644-8. doi: 10.1016/j.mib.2007.09.010. Epub 2007 Nov 5. Curr Opin Microbiol. 2007. PMID: 17981495 Free PMC article. Review.
Cited by
-
Pseudomonas biofilm matrix composition and niche biology.FEMS Microbiol Rev. 2012 Jul;36(4):893-916. doi: 10.1111/j.1574-6976.2011.00322.x. Epub 2012 Jan 23. FEMS Microbiol Rev. 2012. PMID: 22212072 Free PMC article. Review.
-
Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes.Proc Natl Acad Sci U S A. 2006 Jan 3;103(1):171-6. doi: 10.1073/pnas.0507407103. Epub 2005 Dec 22. Proc Natl Acad Sci U S A. 2006. PMID: 16373506 Free PMC article.
-
Catheter-associated urinary tract infection by Pseudomonas aeruginosa is mediated by exopolysaccharide-independent biofilms.Infect Immun. 2014 May;82(5):2048-58. doi: 10.1128/IAI.01652-14. Epub 2014 Mar 4. Infect Immun. 2014. PMID: 24595142 Free PMC article.
-
Strategy to combat biofilms: a focus on biofilm dispersal enzymes.NPJ Biofilms Microbiomes. 2023 Sep 7;9(1):63. doi: 10.1038/s41522-023-00427-y. NPJ Biofilms Microbiomes. 2023. PMID: 37679355 Free PMC article. Review.
-
Inhibition of Pseudomonas aeruginosa Biofilm Formation by Traditional Chinese Medicinal Herb Herba patriniae.Biomed Res Int. 2017;2017:9584703. doi: 10.1155/2017/9584703. Epub 2017 Mar 9. Biomed Res Int. 2017. PMID: 28377931 Free PMC article.
References
-
- Baynham, P. J., and D. J. Wozniak. 1996. Identification and characterization of AlgZ, an AlgT-dependent DNA binding protein required for Pseudomonas aeruginosa algD transcription. Mol. Microbiol. 22:97-108. - PubMed
-
- Christensen, B., C. Sternberg, J. B. Andersen, R. J. Palmer, A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools to study biofilm physiology. Methods Enzymol. 310:20-42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

