Flexibility in the receptor-binding domain of the enzymatic colicin E9 is required for toxicity against Escherichia coli cells
- PMID: 15231784
- PMCID: PMC438598
- DOI: 10.1128/JB.186.14.4520-4527.2004
Flexibility in the receptor-binding domain of the enzymatic colicin E9 is required for toxicity against Escherichia coli cells
Abstract
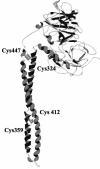
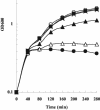
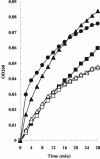
The events that occur after the binding of the enzymatic E colicins to Escherichia coli BtuB receptors that lead to translocation of the cytotoxic domain into the periplasmic space and, ultimately, cell killing are poorly understood. It has been suggested that unfolding of the coiled-coil BtuB receptor binding domain of the E colicins may be an essential step that leads to the loss of immunity protein from the colicin and immunity protein complex and then triggers the events of translocation. We introduced pairs of cysteine mutations into the receptor binding domain of colicin E9 (ColE9) that resulted in the formation of a disulfide bond located near the middle or the top of the R domain. After dithiothreitol reduction, the ColE9 protein with the mutations L359C and F412C (ColE9 L359C-F412C) and the ColE9 protein with the mutations Y324C and L447C (ColE9 Y324C-L447C) were slightly less active than equivalent concentrations of ColE9. On oxidation with diamide, no significant biological activity was seen with the ColE9 L359C-F412C and the ColE9 Y324C-L447C mutant proteins; however diamide had no effect on the activity of ColE9. The presence of a disulfide bond was confirmed in both of the oxidized, mutant proteins by matrix-assisted laser desorption ionization-time of flight mass spectrometry. The loss of biological activity of the disulfide-containing mutant proteins was not due to an indirect effect on the properties of the translocation or DNase domains of the mutant colicins. The data are consistent with a requirement for the flexibility of the coiled-coil R domain after binding to BtuB.
Copyright 2004 American Society for Microbiology
Figures

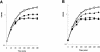

Similar articles
-
Investigating early events in receptor binding and translocation of colicin E9 using synchronized cell killing and proteolytic cleavage.J Bacteriol. 2008 Jun;190(12):4342-50. doi: 10.1128/JB.00047-08. Epub 2008 Apr 11. J Bacteriol. 2008. PMID: 18408035 Free PMC article.
-
Primary events in the colicin translocon: FRET analysis of colicin unfolding initiated by binding to BtuB and OmpF.Biochemistry. 2008 Dec 2;47(48):12802-9. doi: 10.1021/bi800865h. Biochemistry. 2008. PMID: 18986168
-
Structural dynamics of the membrane translocation domain of colicin E9 and its interaction with TolB.J Mol Biol. 2002 May 3;318(3):787-804. doi: 10.1016/S0022-2836(02)00036-0. J Mol Biol. 2002. PMID: 12054823
-
Colicin translocation across the Escherichia coli outer membrane.Biochem Soc Trans. 2012 Dec 1;40(6):1475-9. doi: 10.1042/BST20120255. Biochem Soc Trans. 2012. PMID: 23176501 Review.
-
Killing of E coli cells by E group nuclease colicins.Biochimie. 2002 May-Jun;84(5-6):381-9. doi: 10.1016/s0300-9084(02)01450-5. Biochimie. 2002. PMID: 12423781 Review.
Cited by
-
Mannose-Presenting "Glyco-Colicins" Convert the Bacterial Cell Surface into a Multivalent Adsorption Site for Adherent Bacteria.JACS Au. 2024 Jun 12;4(6):2122-2129. doi: 10.1021/jacsau.4c00365. eCollection 2024 Jun 24. JACS Au. 2024. PMID: 38938796 Free PMC article.
-
Cell entry mechanism of enzymatic bacterial colicins: porin recruitment and the thermodynamics of receptor binding.Proc Natl Acad Sci U S A. 2005 Sep 27;102(39):13849-54. doi: 10.1073/pnas.0503567102. Epub 2005 Sep 15. Proc Natl Acad Sci U S A. 2005. PMID: 16166265 Free PMC article.
-
Comparative molecular dynamics study of the receptor-binding domains in SARS-CoV-2 and SARS-CoV and the effects of mutations on the binding affinity.J Biomol Struct Dyn. 2022 Jul;40(10):4662-4681. doi: 10.1080/07391102.2020.1860829. Epub 2020 Dec 17. J Biomol Struct Dyn. 2022. PMID: 33331243 Free PMC article.
-
Genetic Dissection of the Signaling Cascade that Controls Activation of the Shigella Type III Secretion System from the Needle Tip.Sci Rep. 2016 Jun 9;6:27649. doi: 10.1038/srep27649. Sci Rep. 2016. PMID: 27277624 Free PMC article.
-
Interactions of TolB with the translocation domain of colicin E9 require an extended TolB box.J Bacteriol. 2005 Oct;187(19):6733-41. doi: 10.1128/JB.187.19.6733-6741.2005. J Bacteriol. 2005. PMID: 16166536 Free PMC article.
References
-
- Bénédetti, H., M. Frenette, D. Baty, R. Lloubès, V. Géli, and C. Lazdunski. 1989. Comparison of the uptake systems for the entry of various BtuB group colicins into Escherichia coli. J. Gen. Microbiol. 135:3413-3420. - PubMed
-
- Boetzel, R., E. S. Collins, N. J. Clayden, C. Kleanthous, R. James, and G. R. Moore. 2002. Structural dynamics of the receptor-binding domain of colicin E9. Faraday Discuss. 122:145-162. - PubMed
-
- Bouveret, E., L. Journet, A. Walburger, E. Cascales, H. Bénédetti, and R. Lloubès. 2002. Analysis of the Escherichia coli Tol-Pal and TonB systems by periplasmic production of Tol, TonB, colicin, or phage capsid soluble domains. Biochimie 84:413-421. - PubMed
-
- Bouveret, E., A. Rigal, C. Lazdunski, and H. Bénédetti. 1998. Distinct regions of the colicin A translocation domain are involved in the interaction with TolA and TolB proteins upon import into Escherichia coli. Mol. Microbiol. 27:143-157. - PubMed
-
- Bouveret, E., A. Rigal, C. Lazdunski, and H. Bénédetti. 1997. The N-terminal domain of colicin E3 interacts with TolB which is involved in the colicin translocation step. Mol. Microbiol. 23:909-920. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

