The Bacillus subtilis yqjI gene encodes the NADP+-dependent 6-P-gluconate dehydrogenase in the pentose phosphate pathway
- PMID: 15231785
- PMCID: PMC438568
- DOI: 10.1128/JB.186.14.4528-4534.2004
The Bacillus subtilis yqjI gene encodes the NADP+-dependent 6-P-gluconate dehydrogenase in the pentose phosphate pathway
Abstract
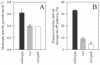
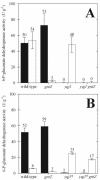
Despite the importance of the oxidative pentose phosphate (PP) pathway as a major source of reducing power and metabolic intermediates for biosynthetic processes, almost no direct genetic or biochemical evidence is available for Bacillus subtilis. Using a combination of knockout mutations in known and putative genes of the oxidative PP pathway and 13C-labeling experiments, we demonstrated that yqjI encodes the NADP+-dependent 6-P-gluconate dehydrogenase, as was hypothesized previously from sequence similarities. Moreover, YqjI was the predominant isoenzyme during glucose and gluconate catabolism, and its role in the oxidative PP pathway could not be played by either of two homologues, GntZ and YqeC. This conclusion is in contrast to the generally held view that GntZ is the relevant isoform; hence, we propose a new designation for yqjI, gndA, the monocistronic gene encoding the principal 6-P-gluconate dehydrogenase. Although we demonstrated the NAD+-dependent 6-P-gluconate dehydrogenase activity of GntZ, gntZ mutants exhibited no detectable phenotype on glucose, and GntZ did not contribute to PP pathway fluxes during growth on glucose. Since gntZ mutants grew normally on gluconate, the functional role of GntZ remains obscure, as does the role of the third homologue, YqeC. Knockout of the glucose-6-P dehydrogenase-encoding zwf gene was primarily compensated for by increased glycolytic fluxes, but about 5% of the catabolic flux was rerouted through the gluconate bypass with glucose dehydrogenase as the key enzyme.
Copyright 2004 American Society for Microbiology
Figures


Similar articles
-
Global metabolic response of Escherichia coli to gnd or zwf gene-knockout, based on 13C-labeling experiments and the measurement of enzyme activities.Appl Microbiol Biotechnol. 2004 Mar;64(1):91-8. doi: 10.1007/s00253-003-1458-5. Epub 2003 Dec 6. Appl Microbiol Biotechnol. 2004. PMID: 14661115
-
Characterization of enzymes involved in the central metabolism of Gluconobacter oxydans.Appl Microbiol Biotechnol. 2010 Oct;88(3):711-8. doi: 10.1007/s00253-010-2779-9. Epub 2010 Jul 30. Appl Microbiol Biotechnol. 2010. PMID: 20676631
-
Enhancement of L-ornithine production by disruption of three genes encoding putative oxidoreductases in Corynebacterium glutamicum.J Ind Microbiol Biotechnol. 2014 Mar;41(3):573-8. doi: 10.1007/s10295-013-1398-8. Epub 2014 Jan 9. J Ind Microbiol Biotechnol. 2014. PMID: 24402505
-
Pathway analysis and metabolic engineering in Corynebacterium glutamicum.Biol Chem. 2000 Sep-Oct;381(9-10):899-910. doi: 10.1515/BC.2000.111. Biol Chem. 2000. PMID: 11076021 Review.
-
[Research progress in glucose-6-phosphate dehydrogenase in higher plants].Sheng Wu Gong Cheng Xue Bao. 2012 Jul;28(7):800-12. Sheng Wu Gong Cheng Xue Bao. 2012. PMID: 23167192 Review. Chinese.
Cited by
-
NADPH-generating systems in bacteria and archaea.Front Microbiol. 2015 Jul 29;6:742. doi: 10.3389/fmicb.2015.00742. eCollection 2015. Front Microbiol. 2015. PMID: 26284036 Free PMC article. Review.
-
YtsJ has the major physiological role of the four paralogous malic enzyme isoforms in Bacillus subtilis.J Bacteriol. 2006 Jul;188(13):4727-36. doi: 10.1128/JB.00167-06. J Bacteriol. 2006. PMID: 16788182 Free PMC article.
-
The metabolic regulation of sporulation and parasporal crystal formation in Bacillus thuringiensis revealed by transcriptomics and proteomics.Mol Cell Proteomics. 2013 May;12(5):1363-76. doi: 10.1074/mcp.M112.023986. Epub 2013 Feb 12. Mol Cell Proteomics. 2013. PMID: 23408684 Free PMC article.
-
Physiological and transcriptomic analyses to reveal underlying phenolic acid action in consecutive monoculture problem of Polygonatum odoratum.BMC Plant Biol. 2021 Aug 7;21(1):362. doi: 10.1186/s12870-021-03135-x. BMC Plant Biol. 2021. PMID: 34364388 Free PMC article.
-
The molecular timeline of a reviving bacterial spore.Mol Cell. 2015 Feb 19;57(4):695-707. doi: 10.1016/j.molcel.2014.12.019. Epub 2015 Feb 5. Mol Cell. 2015. PMID: 25661487 Free PMC article.
References
-
- Adams, M. J., G. H. Ellis, S. Gover, C. E. Naylor, and C. Phillips. 1994. Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism. Structure 2:651-668. - PubMed
-
- Bergmeyer, H. U. 1985. Methods of enzymatic analysis, vol. III. VCH Publishers, Deerfield Beach, Fla.
-
- Blencke, H. M., G. Homuth, H. Ludwig, U. Mader, M. Hecker, and J. Stülke. 2003. Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab. Eng. 5:133-149. - PubMed
-
- Chassagnole, C., N. Noisommit-Rizzi, J. W. Schmid, K. Mauch, and M. Reuss. 2002. Dynamic modeling of the central carbon metabolism of Escherichia coli. Biotechnol. Bioeng. 79:53-73. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

