Phenylphosphate carboxylase: a new C-C lyase involved in anaerobic phenol metabolism in Thauera aromatica
- PMID: 15231788
- PMCID: PMC438602
- DOI: 10.1128/JB.186.14.4556-4567.2004
Phenylphosphate carboxylase: a new C-C lyase involved in anaerobic phenol metabolism in Thauera aromatica
Abstract
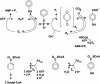
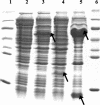
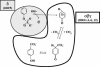
The anaerobic metabolism of phenol in the beta-proteobacterium Thauera aromatica proceeds via carboxylation to 4-hydroxybenzoate and is initiated by the ATP-dependent conversion of phenol to phenylphosphate. The subsequent para carboxylation of phenylphosphate to 4-hydroxybenzoate is catalyzed by phenylphosphate carboxylase, which was purified and studied. This enzyme consists of four proteins with molecular masses of 54, 53, 18, and 10 kDa, whose genes are located adjacent to each other in the phenol gene cluster which codes for phenol-induced proteins. Three of the subunits (54, 53, and 10 kDa) were sufficient to catalyze the exchange of 14CO2 and the carboxyl group of 4-hydroxybenzoate but not phenylphosphate carboxylation. Phenylphosphate carboxylation was restored when the 18-kDa subunit was added. The following reaction model is proposed. The 14CO2 exchange reaction catalyzed by the three subunits of the core enzyme requires the fully reversible release of CO2 from 4-hydroxybenzoate with formation of a tightly enzyme-bound phenolate intermediate. Carboxylation of phenylphosphate requires in addition the 18-kDa subunit, which is thought to form the same enzyme-bound energized phenolate intermediate from phenylphosphate with virtually irreversible release of phosphate. The 54- and 53-kDa subunits show similarity to UbiD of Escherichia coli, which catalyzes the decarboxylation of a 4-hydroxybenzoate derivative in ubiquinone (ubi) biosynthesis. They also show similarity to components of various decarboxylases acting on aromatic carboxylic acids, such as 4-hydroxybenzoate or vanillate, whereas the 10-kDa subunit is unique. The 18-kDa subunit belongs to a hydratase/phosphatase protein family. Phenylphosphate carboxylase is a member of a new family of carboxylases/decarboxylases that act on phenolic compounds, use CO2 as a substrate, do not contain biotin or thiamine diphosphate, require K+ and a divalent metal cation (Mg2+or Mn2+) for activity, and are strongly inhibited by oxygen.
Copyright 2004 American Society for Microbiology
Figures

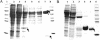


Similar articles
-
Phenylphosphate synthase: a new phosphotransferase catalyzing the first step in anaerobic phenol metabolism in Thauera aromatica.J Bacteriol. 2004 Dec;186(23):8044-57. doi: 10.1128/JB.186.23.8044-8057.2004. J Bacteriol. 2004. PMID: 15547277 Free PMC article.
-
Anaerobic metabolism of phenol in proteobacteria and further studies of phenylphosphate carboxylase.Arch Microbiol. 2009 Dec;191(12):869-78. doi: 10.1007/s00203-009-0519-2. Epub 2009 Oct 17. Arch Microbiol. 2009. PMID: 19838679
-
Carboxylation of phenylphosphate by phenol carboxylase, an enzyme system of anaerobic phenol metabolism.J Bacteriol. 1992 Jun;174(11):3629-36. doi: 10.1128/jb.174.11.3629-3636.1992. J Bacteriol. 1992. PMID: 1592817 Free PMC article.
-
Unusual reactions involved in anaerobic metabolism of phenolic compounds.Biol Chem. 2005 Oct;386(10):989-97. doi: 10.1515/BC.2005.115. Biol Chem. 2005. PMID: 16218871 Review.
-
Activation of Acetone and Other Simple Ketones in Anaerobic Bacteria.J Mol Microbiol Biotechnol. 2016;26(1-3):152-64. doi: 10.1159/000441500. Epub 2016 Mar 10. J Mol Microbiol Biotechnol. 2016. PMID: 26958851 Review.
Cited by
-
Naphthalene Carboxylation in the Sulfate-Reducing Enrichment Culture N47 Is Proposed to Proceed via 1,3-Dipolar Cycloaddition to the Cofactor Prenylated Flavin Mononucleotide.Appl Environ Microbiol. 2023 Mar 29;89(3):e0192722. doi: 10.1128/aem.01927-22. Epub 2023 Feb 23. Appl Environ Microbiol. 2023. PMID: 36815794 Free PMC article.
-
Degradation of phenol via phenylphosphate and carboxylation to 4-hydroxybenzoate by a newly isolated strain of the sulfate-reducing bacterium Desulfobacterium anilini.Appl Environ Microbiol. 2009 Jul;75(13):4248-53. doi: 10.1128/AEM.00203-09. Epub 2009 May 1. Appl Environ Microbiol. 2009. PMID: 19411421 Free PMC article.
-
Genomic and microarray analysis of aromatics degradation in Geobacter metallireducens and comparison to a Geobacter isolate from a contaminated field site.BMC Genomics. 2007 Jun 19;8:180. doi: 10.1186/1471-2164-8-180. BMC Genomics. 2007. PMID: 17578578 Free PMC article.
-
The Catabolic System of Acetovanillone and Acetosyringone in Sphingobium sp. Strain SYK-6 Useful for Upgrading Aromatic Compounds Obtained through Chemical Lignin Depolymerization.Appl Environ Microbiol. 2022 Aug 23;88(16):e0072422. doi: 10.1128/aem.00724-22. Epub 2022 Aug 8. Appl Environ Microbiol. 2022. PMID: 35938864 Free PMC article.
-
Phosphorylation of phenol by phenylphosphate synthase: role of histidine phosphate in catalysis.J Bacteriol. 2006 Nov;188(22):7815-22. doi: 10.1128/JB.00785-06. Epub 2006 Sep 15. J Bacteriol. 2006. PMID: 16980461 Free PMC article.
References
-
- Anders, H. J., A. Kaetzke, P. Kämpfer, W. Ludwig, and G. Fuchs. 1995. Taxonomic position of aromatic-degrading denitrifying pseudomonad strains K 172 and KB 740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Int. J. Sys. Bacteriol. 45327-333. - PubMed
-
- Aoki, K., T. Uemori, R. Shinke, and H. Nishira. 1985. Further characterization of bacterial production of anthranilic acid from aniline. Agric. Biol. Chem. 491151-1158.
-
- Aresta, M., and A. Dibenedetto. 2002. Development of environmentally friendly syntheses: use of enzymes and biomimetic systems for the direct carboxylation of organic substrates. J. Biotechnol. 90113-128. - PubMed
-
- Bak, F., and F. Widdel. 1986. Anaerobic degradation of phenol and phenol derivatives by Desulfobacterium phenolicum sp. nov. Arch. Microbiol. 146177-180.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

