Bacillus subtilis YdiH is a direct negative regulator of the cydABCD operon
- PMID: 15231791
- PMCID: PMC438603
- DOI: 10.1128/JB.186.14.4585-4595.2004
Bacillus subtilis YdiH is a direct negative regulator of the cydABCD operon
Abstract
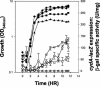
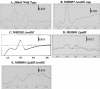
During aerobic respiration, Bacillus subtilis utilizes three terminal oxidases, cytochromes aa3, caa3, and bd. Cytochrome bd is encoded by the cydABCD operon. We report here the first identification of a regulator for the cydABCD operon, YdiH. While working with DeltaresDE mutant strains, we identified colonies which contained suppressor mutations (cmp) which bypassed the requirement for ResD for all phenotypes not associated with cytochrome aa3 or caa3. Mapping identified a class of Tn10 insertions which were close to the cmp locus (Tn10-2) and a second class (Tn10-1) which was inserted in cydD, a gene which appears to be essential to the cmp phenotype. Sequencing of the cmp loci from four independent DeltaresDE cmp isolates yielded four loss-of-function alleles of ydiH, a gene encoding a protein with homology to AT-rich DNA-binding proteins. Additionally, we determined that cytochrome bd was aberrantly expressed in the DeltaresDE cmp background. Together these data led to the hypothesis that YdiH serves as a negative regulator of cydABCD expression, a hypothesis supported by both gel-shift and DNase I footprinting analyses. YdiH protected the cydA promoter region at three 22-bp repeats located in the long 5' untranslated region (193 bp). Induction of the cydABCD operon in a DeltaresDE background showed that expression of the terminal oxidase bd was responsible for the bypass phenotype observed in a DeltaresDE cmp strain, indicating that cytochrome bd expression complemented the loss of cytochromes aa3 and caa3 in the DeltaresDE strain.
Copyright 2004 American Society for Microbiology
Figures

References
-
- Bisschop, A., and W. N. Konings. 1976. Reconstitution of reduced nicotinamide adenine dinucleotide oxidase activity with menadione in membrane vesicles from the menaquinone-deficient Bacillus subtilis aroD. Relation between electron transfer and active transport. Eur. J. Biochem. 67:357-365. - PubMed
-
- Chary, V. K., E. I. Amaya, and P. J. Piggot. 1997. Neomycin- and spectinomycin-resistance replacement vectors for Bacillus subtilis. FEMS Microbiol. Lett. 153:135-139. - PubMed
-
- Cutting, S. M., and P. B. Vander Horn. 1990. Genetic analysis, p. 24-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Inc., New York, N.Y.
-
- de Vrij, W., B. van den Burg, and W. N. Konings. 1987. Spectral and potentiometric analysis of cytochromes from Bacillus subtilis. Eur. J. Biochem. 166:589-595. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

