Structure and electrophysiological properties of the YscC secretin from the type III secretion system of Yersinia enterocolitica
- PMID: 15231798
- PMCID: PMC438636
- DOI: 10.1128/JB.186.14.4645-4654.2004
Structure and electrophysiological properties of the YscC secretin from the type III secretion system of Yersinia enterocolitica
Abstract
YscC is the integral outer membrane component of the type III protein secretion machinery of Yersinia enterocolitica and belongs to the family of secretins. This group of proteins forms stable ring-like oligomers in the outer membrane, which are thought to function as transport channels for macromolecules. The YscC oligomer was purified after solubilization from the membrane with a nonionic detergent. Sodium dodecyl sulfate did not dissociate the oligomer, but it caused a change in electrophoretic mobility and an increase in protease susceptibility, indicating partial denaturation of the subunits within the oligomer. The mass of the homo-oligomer, as determined by scanning transmission electron microscopy, was approximately 1 MDa. Analysis of the angular power spectrum from averaged top views of negatively stained YscC oligomers revealed a 13-fold angular order, suggesting that the oligomer consists of 13 subunits. Reconstituted in planar lipid bilayers, the YscC oligomer displayed a constant voltage-independent conductance of approximately 3 nS, thus forming a stable pore. However, in vivo, the expression of YscC did not lead to an increased permeability of the outer membrane. Electron microscopy revealed that the YscC oligomer is composed of three domains, two stacked rings attached to a conical domain. This structure is consistent with the notion that the secretin forms the upper part of the basal body of the needle structure of the type III secreton.
Copyright 2004 American Society for Microbiology
Figures





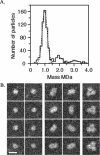

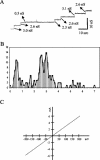
References
-
- Bitter, W., M. Koster, M. Latijnhouwers, H. de Cock, and J. Tommassen. 1998. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol. 27:209-219. - PubMed
-
- Blocker, A., N. Jouihri, E. Larquet, P. Gounon, F. Ebel, C. Parsot, P. Sansonetti, and A. Allaoui. 2001. Structure and composition of the Shigella flexneri “needle complex,” a part of its type III secreton. Mol. Microbiol. 39:652-663. - PubMed
-
- Brok, R., P. Van Gelder, M. Winterhalter, U. Ziese, A. J. Koster, H. de Cock, M. Koster, J. Tommassen, and W. Bitter. 1999. The C-terminal domain of the Pseudomonas secretin XcpQ forms oligomeric rings with pore activity. J. Mol. Biol. 294:1169-1179. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

