Reverse gyrase is not a prerequisite for hyperthermophilic life
- PMID: 15231817
- PMCID: PMC438624
- DOI: 10.1128/JB.186.14.4829-4833.2004
Reverse gyrase is not a prerequisite for hyperthermophilic life
Abstract
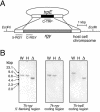
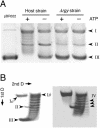
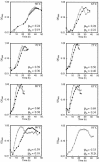
We disrupted the reverse gyrase gene from a hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1. An apparent positive supercoiling activity that was observed in the host strain was not found in the disruptant strain. We found that a lack of reverse gyrase led to a retardation in growth that was more striking at higher temperatures. However, the disruption of the reverse gyrase gene did not lead to a lethal phenotype at 90 degrees C. This study provides experimental evidence that reverse gyrase is not a prerequisite for hyperthermophilic life.
Copyright 2004 American Society for Microbiology
Figures
References
-
- Atomi, H., T. Fukui, T. Kanai, M. Morikawa, and T. Imanaka. 16 April 2004, posting date. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea [Online.] http://archaea.ws/archive/pdf/volume1/issue4/1-Imanaka.pdf. - PMC - PubMed
-
- Blöchl, E., R. Rachel, S. Burggraf, D. Hafenbradl, H. W. Jannasch, and K. O. Stetter. 1997. Pyrolobus fumarii, gen. and sp. nov., represents a novel group of archaea, extending the upper temperature limit for life to 113°C. Extremophiles 1:14-21. - PubMed
-
- Forterre, P. 2002. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 18:236-237. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

