Synaptotagmin gene content of the sequenced genomes
- PMID: 15238157
- PMCID: PMC471550
- DOI: 10.1186/1471-2164-5-43
Synaptotagmin gene content of the sequenced genomes
Abstract
Background: Synaptotagmins exist as a large gene family in mammals. There is much interest in the function of certain family members which act crucially in the regulated synaptic vesicle exocytosis required for efficient neurotransmission. Knowledge of the functions of other family members is relatively poor and the presence of Synaptotagmin genes in plants indicates a role for the family as a whole which is wider than neurotransmission. Identification of the Synaptotagmin genes within completely sequenced genomes can provide the entire Synaptotagmin gene complement of each sequenced organism. Defining the detailed structures of all the Synaptotagmin genes and their encoded products can provide a useful resource for functional studies and a deeper understanding of the evolution of the gene family. The current rapid increase in the number of sequenced genomes from different branches of the tree of life, together with the public deposition of evolutionarily diverse transcript sequences make such studies worthwhile.
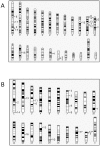
Results: I have compiled a detailed list of the Synaptotagmin genes of Caenorhabditis, Anopheles, Drosophila, Ciona, Danio, Fugu, Mus, Homo, Arabidopsis and Oryza by examining genomic and transcript sequences from public sequence databases together with some transcript sequences obtained by cDNA library screening and RT-PCR. I have compared all of the genes and investigated the relationship between plant Synaptotagmins and their non-Synaptotagmin counterparts.
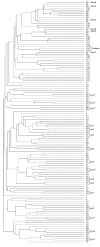
Conclusions: I have identified and compared 98 Synaptotagmin genes from 10 sequenced genomes. Detailed comparison of transcript sequences reveals abundant and complex variation in Synaptotagmin gene expression and indicates the presence of Synaptotagmin genes in all animals and land plants. Amino acid sequence comparisons indicate patterns of conservation and diversity in function. Phylogenetic analysis shows the origin of Synaptotagmins in multicellular eukaryotes and their great diversification in animals. Synaptotagmins occur in land plants and animals in combinations of 4-16 in different species. The detailed delineation of the Synaptotagmin genes presented here, will allow easier identification of Synaptotagmins in future. Since the functional roles of many of these genes are unknown, this gene collection provides a useful resource for future studies.
Figures








References
-
- Geppert M, Archer BT, III, Südhof TC. Synaptotagmin II a novel differentially distributed form of synaptotagmin. J Biol Chem. 1991;266:13548–13552. - PubMed
-
- Mizuta M, Inagaki N, Nemoto Y, Matsukura S, Takahashi M, Seino S. Synaptotagmin III is a novel isoform of rat synaptotagmin expressed in endocrine and neuronal cells. J Biol Chem. 1994;269:11675–11678. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

