Biologically meaningful expression profiling across species using heterologous hybridization to a cDNA microarray
- PMID: 15238158
- PMCID: PMC471549
- DOI: 10.1186/1471-2164-5-42
Biologically meaningful expression profiling across species using heterologous hybridization to a cDNA microarray
Abstract
Background: Unravelling the path from genotype to phenotype, as it is influenced by an organism's environment, is one of the central goals in biology. Gene expression profiling by means of microarrays has become very prominent in this endeavour, although resources exist only for relatively few model systems. As genomics has matured into a comparative research program, expression profiling now also provides a powerful tool for non-traditional model systems to elucidate the molecular basis of complex traits.
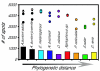
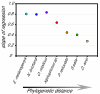
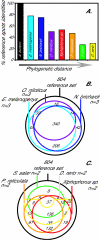
Results: Here we present a microarray constructed with approximately 4500 features, derived from a brain-specific cDNA library for the African cichlid fish Astatotilapia burtoni (Perciformes). Heterologous hybridization, targeting RNA to an array constructed for a different species, is used for eight different fish species. We quantified the concordance in gene expression profiles across these species (number of genes and fold-changes). Although most robust when target RNA is derived from closely related species (<10 MA divergence time), our results showed consistent profiles for other closely related taxa (approximately 65 MA divergence time) and, to a lesser extent, even very distantly related species (>200 MA divergence time).
Conclusion: This strategy overcomes some of the restrictions imposed on model systems that are of importance for evolutionary and ecological studies, but for which only limited sequence information is available. Our work validates the use of expression profiling for functional genomics within a comparative framework and provides a foundation for the molecular and cellular analysis of complex traits in a wide range of organisms.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

