RAG: RNA-As-Graphs web resource
- PMID: 15238163
- PMCID: PMC471545
- DOI: 10.1186/1471-2105-5-88
RAG: RNA-As-Graphs web resource
Abstract
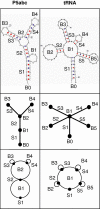
Background: The proliferation of structural and functional studies of RNA has revealed an increasing range of RNA's structural repertoire. Toward the objective of systematic cataloguing of RNA's structural repertoire, we have recently described the basis of a graphical approach for organizing RNA secondary structures, including existing and hypothetical motifs.
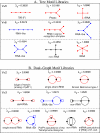
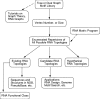
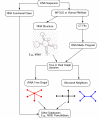
Description: We now present an RNA motif database based on graph theory, termed RAG for RNA-As-Graphs, to catalogue and rank all theoretically possible, including existing, candidate and hypothetical, RNA secondary motifs. The candidate motifs are predicted using a clustering algorithm that classifies RNA graphs into RNA-like and non-RNA groups. All RNA motifs are filed according to their graph vertex number (RNA length) and ranked by topological complexity.
Conclusions: RAG's quantitative cataloguing allows facile retrieval of all classes of RNA secondary motifs, assists identification of structural and functional properties of user-supplied RNA sequences, and helps stimulate the search for novel RNAs based on predicted candidate motifs.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

