Evolutionary conservation and selection of human disease gene orthologs in the rat and mouse genomes
- PMID: 15239832
- PMCID: PMC463309
- DOI: 10.1186/gb-2004-5-7-r47
Evolutionary conservation and selection of human disease gene orthologs in the rat and mouse genomes
Abstract
Background: Model organisms have contributed substantially to our understanding of the etiology of human disease as well as having assisted with the development of new treatment modalities. The availability of the human, mouse and, most recently, the rat genome sequences now permit the comprehensive investigation of the rodent orthologs of genes associated with human disease. Here, we investigate whether human disease genes differ significantly from their rodent orthologs with respect to their overall levels of conservation and their rates of evolutionary change.
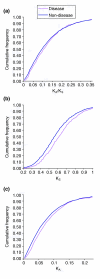
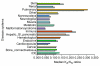
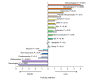
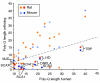
Results: Human disease genes are unevenly distributed among human chromosomes and are highly represented (99.5%) among human-rodent ortholog sets. Differences are revealed in evolutionary conservation and selection between different categories of human disease genes. Although selection appears not to have greatly discriminated between disease and non-disease genes, synonymous substitution rates are significantly higher for disease genes. In neurological and malformation syndrome disease systems, associated genes have evolved slowly whereas genes of the immune, hematological and pulmonary disease systems have changed more rapidly. Amino-acid substitutions associated with human inherited disease occur at sites that are more highly conserved than the average; nevertheless, 15 substituting amino acids associated with human disease were identified as wild-type amino acids in the rat. Rodent orthologs of human trinucleotide repeat-expansion disease genes were found to contain substantially fewer of such repeats. Six human genes that share the same characteristics as triplet repeat-expansion disease-associated genes were identified; although four of these genes are expressed in the brain, none is currently known to be associated with disease.
Conclusions: Most human disease genes have been retained in rodent genomes. Synonymous nucleotide substitutions occur at a higher rate in disease genes, a finding that may reflect increased mutation rates in the chromosomal regions in which disease genes are found. Rodent orthologs associated with neurological function exhibit the greatest evolutionary conservation; this suggests that rodent models of human neurological disease are likely to most faithfully represent human disease processes. However, with regard to neurological triplet repeat expansion-associated human disease genes, the contraction, relative to human, of rodent trinucleotide repeats suggests that rodent loci may not achieve a 'critical repeat threshold' necessary to undergo spontaneous pathological repeat expansions. The identification of six genes in this study that have multiple characteristics associated with repeat expansion-disease genes raises the possibility that not all human loci capable of facilitating neurological disease by repeat expansion have as yet been identified.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

