Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic ocean
- PMID: 15240292
- PMCID: PMC444831
- DOI: 10.1128/AEM.70.7.4129-4135.2004
Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic ocean
Abstract
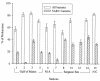
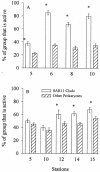
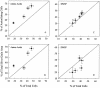
SAR11 bacteria are abundant in marine environments, often accounting for 35% of total prokaryotes in the surface ocean, but little is known about their involvement in marine biogeochemical cycles. Previous studies reported that SAR11 bacteria are very small and potentially have few ribosomes, indicating that SAR11 bacteria could have low metabolic activities and could play a smaller role in the flux of dissolved organic matter than suggested by their abundance. To determine the ecological activity of SAR11 bacteria, we used a combination of microautoradiography and fluorescence in situ hybridization (Micro-FISH) to measure assimilation of (3)H-amino acids and [(35)S]dimethylsulfoniopropionate (DMSP) by SAR11 bacteria in the coastal North Atlantic Ocean and the Sargasso Sea. We found that SAR11 bacteria were often abundant in surface waters, accounting for 25% of all prokaryotes on average. SAR11 bacteria were typically as large as, if not larger than, other prokaryotes. Additionally, more than half of SAR11 bacteria assimilated dissolved amino acids and DMSP, whereas about 40% of other prokaryotes assimilated these compounds. Due to their high abundance and activity, SAR11 bacteria were responsible for about 50% of amino acid assimilation and 30% of DMSP assimilation in surface waters. The contribution of SAR11 bacteria to amino acid assimilation was greater than would be expected based on their overall abundance, implying that SAR11 bacteria outcompete other prokaryotes for these labile compounds. These data suggest that SAR11 bacteria are highly active and play a significant role in C, N, and S cycling in the ocean.
Figures


References
-
- Charlson, R. J., J. E. Lovelock, M. O. Andreae, and S. G. Warren. 1987. Oceanic phytoplankton, atmospheric sulfur, cloud albedo and climate. Nature 326:655-661.
-
- Cottrell, M. T., and D. L. Kirchman. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48:168-178.
-
- Elser, J. J., L. B. Stabler, and R. P. Hassett. 1995. Nutrient limitation of bacterial growth and rates of bacterivory in lakes and oceans—a comparative study. Aquat. Microb. Ecol. 9:105-110.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

