Molecular analysis of shower curtain biofilm microbes
- PMID: 15240300
- PMCID: PMC444822
- DOI: 10.1128/AEM.70.7.4187-4192.2004
Molecular analysis of shower curtain biofilm microbes
Abstract
Households provide environments that encourage the formation of microbial communities, often as biofilms. Such biofilms constitute potential reservoirs for pathogens, particularly for immune-compromised individuals. One household environment that potentially accumulates microbial biofilms is that provided by vinyl shower curtains. Over time, vinyl shower curtains accumulate films, commonly referred to as "soap scum," which microscopy reveals are constituted of lush microbial biofilms. To determine the kinds of microbes that constitute shower curtain biofilms and thereby to identify potential opportunistic pathogens, we conducted an analysis of rRNA genes obtained by PCR from four vinyl shower curtains from different households. Each of the shower curtain communities was highly complex. No sequence was identical to one in the databases, and no identical sequences were encountered in the different communities. However, the sequences generally represented similar phylogenetic kinds of organisms. Particularly abundant sequences represented members of the alpha-group of proteobacteria, mainly Sphingomonas spp. and Methylobacterium spp. Both of these genera are known to include opportunistic pathogens, and several of the sequences obtained from the environmental DNA samples were closely related to known pathogens. Such organisms have also been linked to biofilm formation associated with water reservoirs and conduits. In addition, the study detected many other kinds of organisms at lower abundances. These results show that shower curtains are a potential source of opportunistic pathogens associated with biofilms. Frequent cleaning or disposal of shower curtains is indicated, particularly in households with immune-compromised individuals.
Figures
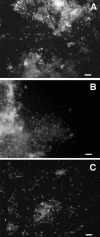

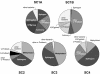
References
-
- Berrocal, A. M., I. U. Scott, D. Miller, and H. W. Flynn, Jr. 2002. Endophthalmitis caused by Moraxella osloensis. Graefe's Arch. Clin. Exp. Ophthalmol. 240:329-330. - PubMed
-
- Boe-Hansen, R., H. J. Albrechtsen, E. Arvin, and C. Jorgensen. 2002. Bulk water phase and biofilm growth in drinking water at low nutrient conditions. Water Res. 36:4477-4486. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

