Molecular analysis of carbon monoxide-oxidizing bacteria associated with recent Hawaiian volcanic deposits
- PMID: 15240307
- PMCID: PMC444802
- DOI: 10.1128/AEM.70.7.4242-4248.2004
Molecular analysis of carbon monoxide-oxidizing bacteria associated with recent Hawaiian volcanic deposits
Abstract
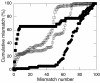
Genomic DNA extracts from four sites at Kilauea Volcano were used as templates for PCR amplification of the large subunit (coxL) of aerobic carbon monoxide dehydrogenase. The sites included a 42-year-old tephra deposit, a 108-year-old lava flow, a 212-year-old partially vegetated ash-and-tephra deposit, and an approximately 300-year-old forest. PCR primers amplified coxL sequences from the OMP clade of CO oxidizers, which includes isolates such as Oligotropha carboxidovorans, Mycobacterium tuberculosis, and Pseudomonas thermocarboxydovorans. PCR products were used to create clone libraries that provide the first insights into the diversity and phylogenetic affiliations of CO oxidizers in situ. On the basis of phylogenetic and statistical analyses, clone libraries for each site were distinct. Although some clone sequences were similar to coxL sequences from known organisms, many sequences appeared to represent phylogenetic lineages not previously known to harbor CO oxidizers. On the basis of average nucleotide diversity and average pairwise difference, a forested site supported the most diverse CO-oxidizing populations, while an 1894 lava flow supported the least diverse populations. Neither parameter correlated with previous estimates of atmospheric CO uptake rates, but both parameters correlated positively with estimates of microbial biomass and respiration. Collectively, the results indicate that the CO oxidizer functional group associated with recent volcanic deposits of the remote Hawaiian Islands contains substantial and previously unsuspected diversity.
Figures

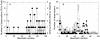

References
-
- Crews, T. E., H. Farrington, and P. M. Vitousek. 2000. Changes in asymbiotic, heterotrophic nitrogen fixation on leaf litter of Metrosideros polymorpha with long-term ecosystem development in Hawaii. Ecosystems 3:386-395.
-
- Dedysh, S. V., M. Derakshani, and W. Liesack. 2001. Detection and enumeration of methanotrophs in acidic Sphagnum peat by 16S rRNA fluorescence in situ hybridization, including the use of newly developed oligonucleotide probes for Methylocella palustris. Appl. Environ. Microbiol. 67:4850-4857. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

