Natural atypical Listeria innocua strains with Listeria monocytogenes pathogenicity island 1 genes
- PMID: 15240309
- PMCID: PMC444784
- DOI: 10.1128/AEM.70.7.4256-4266.2004
Natural atypical Listeria innocua strains with Listeria monocytogenes pathogenicity island 1 genes
Abstract
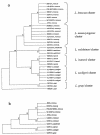
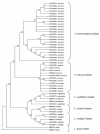
Identification of bona fide Listeria isolates into the six species of the genus normally requires only a few tests. Aberrant isolates do occur, but even then only one or two extra confirmatory tests are generally needed for identification to species level. We have discovered a hemolytic-positive, rhamnose and xylose fermentation-negative Listeria strain with surprising recalcitrance to identification to the species level due to contradictory results in standard confirmatory tests. The issue had to be resolved by using total DNA-DNA hybridization testing and then confirmed by further specific PCR-based tests including a Listeria microarray assay. The results show that this isolate is indeed a novel one. Its discovery provides the first fully documented instance of a hemolytic Listeria innocua strain. This species, by definition, is typically nonhemolytic. The L. innocua isolate contains all the members of the PrfA-regulated virulence gene cluster (Listeria pathogenicity island 1) of L. monocytogenes. It is avirulent in the mouse pathogenicity test. Avirulence is likely at least partly due to the absence of the L. monocytogenes-specific allele of iap, as well as the absence of inlA, inlB, inlC, and daaA. At least two of the virulence cluster genes, hly and plcA, which encode the L. monocytogenes hemolysin (listeriolysin O) and inositol-specific phospholipase C, respectively, are phenotypically expressed in this L. innocua strain. The detection by PCR assays of specific L. innocua genes (lin0198, lin0372, lin0419, lin0558, lin1068, lin1073, lin1074, lin2454, and lin2693) and noncoding intergenic regions (lin0454-lin0455 and nadA-lin2134) in the strain is consistent with its L. innocua DNA-DNA hybridization identity. Additional distinctly different hemolytic L. innocua strains were also studied.
Figures
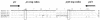
References
-
- Anonymous. 2002. Draft assessment of the relative risk to public health from food borne Listeria monocytogenes among selected categories of ready-to-eat foods. Food and Drug Administration, Center for Food Safety and Applied Nutrition, and U.S. Department of Agriculture, Food Safety and Inspection Service. [Online.] http://www.foodsafety.gov/∼dms/lmrisk.html.
-
- Bennett, R. W., and R. E. Weaver. 2003. Bacteriological analytical manual, chapter 11. Food and Drug Administration. [Online.] http://www.cfsan.fda.gov/∼ebam/bam-11.html.
-
- Cai, S., and M. Wiedmann. 2001. Characterization of the prfA virulence gene cluster insertion site in non-hemolytic Listeria spp.: probing the evolution of the Listeria virulence gene island. Curr. Microbiol. 43:271-277. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

