Quantification of Protein-Lipid Selectivity using FRET: Application to the M13 Major Coat Protein
- PMID: 15240469
- PMCID: PMC1304355
- DOI: 10.1529/biophysj.104.040337
Quantification of Protein-Lipid Selectivity using FRET: Application to the M13 Major Coat Protein
Abstract
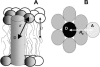
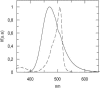
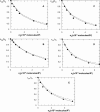
Quantification of lipid selectivity by membrane proteins has been previously addressed mainly from electron spin resonance studies. We present here a new methodology for quantification of protein-lipid selectivity based on fluorescence resonance energy transfer. A mutant of M13 major coat protein was labeled with 7-diethylamino-3((4'iodoacetyl)amino)phenyl-4-methylcoumarin to be used as the donor in energy transfer studies. Phospholipids labeled with N-(7-nitro-2-1,3-benzoxadiazol-4-yl) were selected as the acceptors. The dependence of protein-lipid selectivity on both hydrophobic mismatch and headgroup family was determined. M13 major coat protein exhibited larger selectivity toward phospholipids which allow for a better hydrophobic matching. Increased selectivity was also observed for anionic phospholipids and the relative association constants agreed with the ones already presented in the literature and obtained through electron spin resonance studies. This result led us to conclude that fluorescence resonance energy transfer is a promising methodology in protein-lipid selectivity studies.
Figures

Similar articles
-
Förster resonance energy transfer as a tool for quantification of protein-lipid selectivity.Methods Mol Biol. 2013;974:219-32. doi: 10.1007/978-1-62703-275-9_10. Methods Mol Biol. 2013. PMID: 23404278
-
FRET study of membrane proteins: determination of the tilt and orientation of the N-terminal domain of M13 major coat protein.Biophys J. 2007 Feb 15;92(4):1296-305. doi: 10.1529/biophysj.106.095026. Epub 2006 Nov 17. Biophys J. 2007. PMID: 17114224 Free PMC article.
-
Membrane-bound conformation of M13 major coat protein: a structure validation through FRET-derived constraints.J Biol Chem. 2005 Nov 18;280(46):38522-7. doi: 10.1074/jbc.M505875200. Epub 2005 Sep 8. J Biol Chem. 2005. PMID: 16150733
-
Modeling FRET to investigate the selectivity of lactose permease of Escherichia coli for lipids.Mol Membr Biol. 2014 Jun;31(4):120-30. doi: 10.3109/09687688.2014.915351. Epub 2014 May 15. Mol Membr Biol. 2014. PMID: 24826799 Review.
-
Quantification of protein-lipid selectivity using FRET.Eur Biophys J. 2010 Mar;39(4):565-78. doi: 10.1007/s00249-009-0532-z. Eur Biophys J. 2010. PMID: 20238256 Free PMC article. Review.
Cited by
-
FRET in Membrane Biophysics: An Overview.Front Physiol. 2011 Nov 15;2:82. doi: 10.3389/fphys.2011.00082. eCollection 2011. Front Physiol. 2011. PMID: 22110442 Free PMC article.
-
Lipid bilayer topology of the transmembrane alpha-helix of M13 Major coat protein and bilayer polarity profile by site-directed fluorescence spectroscopy.Biophys J. 2004 Sep;87(3):1445-55. doi: 10.1529/biophysj.104.043208. Biophys J. 2004. PMID: 15345527 Free PMC article.
-
Transmembrane peptides influence the affinity of sterols for phospholipid bilayers.Biophys J. 2010 Jul 21;99(2):526-33. doi: 10.1016/j.bpj.2010.04.052. Biophys J. 2010. PMID: 20643071 Free PMC article.
-
Motional restrictions of membrane proteins: a site-directed spin labeling study.Biophys J. 2006 Nov 1;91(9):3341-8. doi: 10.1529/biophysj.106.090308. Epub 2006 Aug 11. Biophys J. 2006. PMID: 16905615 Free PMC article.
-
Non-uniform membrane probe distribution in resonance energy transfer: application to protein-lipid selectivity.J Fluoresc. 2006 Mar;16(2):161-72. doi: 10.1007/s10895-005-0036-x. Epub 2006 Mar 11. J Fluoresc. 2006. PMID: 16532364
References
-
- Abrams, F. S., and E. London. 1993. Extension of the parallax analysis of membrane penetration depth to the polar region of model membranes: use of fluorescence quenching by a spin-label attached to the phospholipids polar headgroup. Biochemistry. 32:10826–10831. - PubMed
-
- Berberan-Santos, M. N., and M. J. E. Prieto. 1987. Energy transfer in spherical geometry. Application to micelles. J. Chem. Soc. Faraday Trans. 283:1391–1409.
-
- Bonini, I. C., S. S. Antollini, C. Gutiérrez-Merino, and F. J. Barrantes. 2002. Sphingomyelin composition and physical asymmetries in native acetylcholine receptor-rich membranes. Eur. Biophys. J. 31:417–427. - PubMed
-
- Chattopadhyay, A. 1990. Chemistry and biology of N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-labeled lipids: fluorescent probes of biological and model membranes. Chem. Phys. Lipids. 53:1–15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

