The unfolding action of GroEL on a protein substrate
- PMID: 15240489
- PMCID: PMC1304377
- DOI: 10.1529/biophysj.103.037333
The unfolding action of GroEL on a protein substrate
Abstract
A molecular dynamics simulation of the active unfolding of denatured rhodanese by the chaperone GroEL is presented. The compact denatured protein is bound initially to the cis cavity and forms stable contacts with several of the subunits. As the cis ring apical domains of GroEL undergo the transition from the closed to the more open (ATP-bound) state, they exert a force on rhodanese that leads to the increased unfolding of certain loops. The contacts between GroEL and rhodanese are analyzed and their variation during the GroEL transition is shown. The major contacts, which give rise to the stretching force, are found to be similar to those observed in crystal structures of peptides bound to the apical domains. The results of the simulation show that multidomain interactions play an essential role, in accord with experiments. Implications of the results for mutation experiments and for the action of GroEL are discussed.
Figures



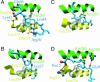
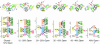


Similar articles
-
On the chaperonin activity of GroEL at heat-shock temperature.Int J Biochem Cell Biol. 2005 Jul;37(7):1375-85. doi: 10.1016/j.biocel.2005.01.007. Int J Biochem Cell Biol. 2005. PMID: 15833270
-
Solution structures of GroEL and its complex with rhodanese from small-angle neutron scattering.Structure. 1996 Jan 15;4(1):79-88. doi: 10.1016/s0969-2126(96)00011-1. Structure. 1996. PMID: 8805508
-
The oligomeric structure of GroEL/GroES is required for biologically significant chaperonin function in protein folding.Nat Struct Biol. 1998 Nov;5(11):977-85. doi: 10.1038/2952. Nat Struct Biol. 1998. PMID: 9808043
-
Molecular chaperone GroEL/ES: unfolding and refolding processes.Biochemistry (Mosc). 2013 Dec;78(13):1405-14. doi: 10.1134/S0006297913130038. Biochemistry (Mosc). 2013. PMID: 24490731 Review.
-
Molecular chaperones: inside and outside the Anfinsen cage.Curr Biol. 2001 Dec 11;11(24):R1038-40. doi: 10.1016/s0960-9822(01)00620-0. Curr Biol. 2001. PMID: 11747844 Review.
Cited by
-
GroEL-mediated protein folding: making the impossible, possible.Crit Rev Biochem Mol Biol. 2006 Jul-Aug;41(4):211-39. doi: 10.1080/10409230600760382. Crit Rev Biochem Mol Biol. 2006. PMID: 16849107 Free PMC article. Review.
-
CHARMM: the biomolecular simulation program.J Comput Chem. 2009 Jul 30;30(10):1545-614. doi: 10.1002/jcc.21287. J Comput Chem. 2009. PMID: 19444816 Free PMC article. Review.
-
Allosteric transitions in the chaperonin GroEL are captured by a dominant normal mode that is most robust to sequence variations.Biophys J. 2007 Oct 1;93(7):2289-99. doi: 10.1529/biophysj.107.105270. Epub 2007 Jun 8. Biophys J. 2007. PMID: 17557788 Free PMC article.
-
Polar or apolar--the role of polarity for urea-induced protein denaturation.PLoS Comput Biol. 2008 Nov;4(11):e1000221. doi: 10.1371/journal.pcbi.1000221. Epub 2008 Nov 14. PLoS Comput Biol. 2008. PMID: 19008937 Free PMC article.
-
Coupling between allosteric transitions in GroEL and assisted folding of a substrate protein.Proc Natl Acad Sci U S A. 2007 May 22;104(21):8803-8. doi: 10.1073/pnas.0700607104. Epub 2007 May 11. Proc Natl Acad Sci U S A. 2007. PMID: 17496143 Free PMC article.
References
-
- Anfinsen, C. 1973. Principles that govern the folding of protein chains. Science. 181:223–230. - PubMed
-
- Betancourt, M., and D. Thirumalai. 1999. Exploring the kinetic requirements for enhancement of protein folding rates in the GroEL cavity. J. Mol. Biol. 287:627–644. - PubMed
-
- Böckmann, R., and H. Grubmüller. 2002. Nanoseconds molecular dynamics simulation of primary mechanical energy transfer steps in F1-ATP synthase. Nat. Struct. Biol. 9:198–202. - PubMed
-
- Boisvert, D., J. Wang, Z. Otwinowski, A. Horwich, and P. Sigler. 1996. The 2.4 Å crystal structure of the bacterial chaperonin GroEL complexed with ATPγS. Nat. Struct. Biol. 3:170–177. - PubMed
-
- Brinker, A., G. Pfeifer, M. Kerner, D. Naylor, F. Hartl, and M. Hayer-Hartl. 2001. Dual function of protein confinement in chaperonin-assisted protein folding. Cell. 107:223–233. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

