Nutritional homeostasis in batch and steady-state culture of yeast
- PMID: 15240820
- PMCID: PMC515343
- DOI: 10.1091/mbc.e04-04-0306
Nutritional homeostasis in batch and steady-state culture of yeast
Abstract
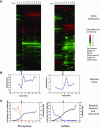
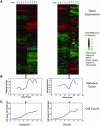
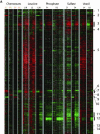
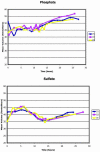
We studied the physiological response to limitation by diverse nutrients in batch and steady-state (chemostat) cultures of S. cerevisiae. We found that the global pattern of transcription in steady-state cultures in limiting phosphate or sulfate is essentially identical to that of batch cultures growing in the same medium just before the limiting nutrient is completely exhausted. The massive stress response and complete arrest of the cell cycle that occurs when nutrients are fully exhausted in batch cultures is not observed in the chemostat, indicating that the cells in the chemostat are "poor, not starving." Similar comparisons using leucine or uracil auxotrophs limited on leucine or uracil again showed patterns of gene expression in steady-state closely resembling those of corresponding batch cultures just before they exhaust the nutrient. Although there is also a strong stress response in the auxotrophic batch cultures, cell cycle arrest, if it occurs at all, is much less uniform. Many of the differences among the patterns of gene expression between the four nutrient limitations are interpretable in light of known involvement of the genes in stress responses or in the regulation or execution of particular metabolic pathways appropriate to the limiting nutrient. We conclude that cells adjust their growth rate to nutrient availability and maintain homeostasis in the same way in batch and steady state conditions; cells in steady-state cultures are in a physiological condition normally encountered in batch cultures.
Figures




References
-
- Boer, V.M., de Winde, J.H., Pronk, J.T., and Piper, M.D. (2003). The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 278, 3265-3274. - PubMed
-
- Carroll, A.S., and O'Shea, E.K. (2002). Pho85 and signaling environmental conditions. Trends Biochem. Sci. 27, 87-93. - PubMed
-
- Chen, P.S., Toribara, T., and Warner, H. (1956). Microdetermination of phosphorus. Anal. Chem. 28, 1756-1758.
-
- Denis-Duphil, M. (1989). Pyrimidine biosynthesis in Saccharomyces cerevisiae: the ura2 cluster gene, its multifunctional enzyme product, and other structural or regulatory genes involved in de novo UMP synthesis. Biochem. Cell Biol. 67, 612-631. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

