New strategy for the representation and the integration of biomolecular knowledge at a cellular scale
- PMID: 15240831
- PMCID: PMC484170
- DOI: 10.1093/nar/gkh681
New strategy for the representation and the integration of biomolecular knowledge at a cellular scale
Abstract
The combination of sequencing and post-sequencing experimental approaches produces huge collections of data that are highly heterogeneous both in structure and in semantics. We propose a new strategy for the integration of such data. This strategy uses structured sets of sequences as a unified representation of biological information and defines a probabilistic measure of similarity between the sets. Sets can be composed of sequences that are known to have a biological relationship (e.g. proteins involved in a complex or a pathway) or that share similar values for a particular attribute (e.g. expression profile). We have developed a software, BlastSets, which implements this strategy. It exploits a database where the sets derived from diverse biological information can be deposited using a standard XML format. For a given query set, BlastSets returns target sets found in the database whose similarity to the query is statistically significant. The tool allowed us to automatically identify verified relationships between correlated expression profiles and biological pathways using publicly available data for Saccharomyces cerevisiae. It was also used to retrieve the members of a complex (ribosome) based on the mining of expression profiles. These first results validate the relevance of the strategy and demonstrate the promising potential of BlastSets.
Figures

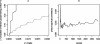

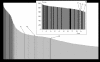
References
-
- Etzold T., Ulyanov,A. and Argos,P. (1996) SRS: information retrieval system for molecular biology data banks. Methods Enzymol., 266, 114–128. - PubMed
-
- Schuler G.D., Epstein,J.A., Ohkawa,H. and Kans,J.A. (1996) Entrez: molecular biology database and retrieval system. Methods Enzymol., 266, 141–162. - PubMed
-
- Tomita M., Hashimoto,K., Takahashi,K., Shimizu,T.S., Matsuzaki,Y., Miyoshi,F., Saito,K., Tanida,S., Yugi,K., Venter,J.C. et al. (1999) E-CELL: software environment for whole-cell simulation. Bioinformatics, 15, 72–84. - PubMed
-
- de Jong H., Geiselmann,J., Hernandez,C. and Page,M. (2003) Genetic Network Analyzer: qualitative simulation of genetic regulatory networks. Bioinformatics, 19, 336–344. - PubMed
-
- Danchin A. (1998) La barque de Delphes—Ce que révèle le texte des génomes. Odile Jacob, Paris, France.

