Insight into the PrPC-->PrPSc conversion from the structures of antibody-bound ovine prion scrapie-susceptibility variants
- PMID: 15240887
- PMCID: PMC478560
- DOI: 10.1073/pnas.0400014101
Insight into the PrPC-->PrPSc conversion from the structures of antibody-bound ovine prion scrapie-susceptibility variants
Abstract
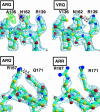
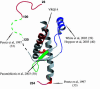
Prion diseases are associated with the conversion of the alpha-helix rich prion protein (PrPC) into a beta-structure-rich insoluble conformer (PrPSc) that is thought to be infectious. The mechanism for the PrPC-->PrPSc conversion and its relationship with the pathological effects of prion diseases are poorly understood, partly because of our limited knowledge of the structure of PrPSc. In particular, the way in which mutations in the PRNP gene yield variants that confer different susceptibilities to disease needs to be clarified. We report here the 2.5-A-resolution crystal structures of three scrapie-susceptibility ovine PrP variants complexed with an antibody that binds to PrPC and to PrPSc; they identify two important features of the PrPC-->PrPSc conversion. First, the epitope of the antibody mainly consists of the last two turns of ovine PrP second alpha-helix. We show that this is a structural invariant in the PrPC-->PrPSc conversion; taken together with biochemical data, this leads to a model of the conformational change in which the two PrPC C-terminal alpha-helices are conserved in PrPSc, whereas secondary structure changes are located in the N-terminal alpha-helix. Second, comparison of the structures of scrapie-sensitivity variants defines local changes in distant parts of the protein that account for the observed differences of PrPC stability, resistant variants being destabilized compared with sensitive ones. Additive contributions of these sensitivity-modulating mutations to resistance suggest a possible causal relationship between scrapie resistance and lowered stability of the PrP protein.
Figures


References
-
- Bueler, H., Aguzzi, A., Sailer, A., Greiner, R. A., Autenried, P., Aguet, M. & Weissmann, C. (1993) Cell 73, 1339–1347. - PubMed
-
- Lasmezas, C. I., Deslys, J. P., Robain, O., Jaegly, A., Beringue, V., Peyrin, J. M., Fournier, J. G., Hauw, J. J., Rossier, J. & Dormont, D. (1997) Science 275, 402–405. - PubMed
-
- Goldmann, W., Martin, T., Foster, J., Hughes, S., Smith, G., Hughes, K., Dawson, M. & Hunter, N. (1996) J. Gen. Virol. 77, 2885–2891. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

