Protein arginine methylation during lytic adenovirus infection
- PMID: 15242333
- PMCID: PMC1134066
- DOI: 10.1042/BJ20040210
Protein arginine methylation during lytic adenovirus infection
Abstract
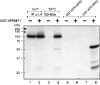
Arginine methylation of proteins affects major processes in the cell, including transcriptional regulation, mRNA metabolism, signal transduction and protein sorting. Arginine methylation of Ad (adenovirus) E1B 55-kDa-associated protein E1B-AP5 was recently described by us [Kzhyshkowska, Schutt, Liss, Kremmer, Stauber, Wolf and Dobner (2001) Biochem. J. 358, 305-314]. In this first example of protein arginine methylation analysis in Ad-infected cells, we investigated methylation of the E1B-AP5 and the viral L4-100 kDa protein. We demonstrate that E1B-AP5 methylation is enhanced during the course of infection in a cell-type-specific manner. We also show that L4-100 kDa is efficiently methylated in Ad-infected cells. L4-100 kDa formed complex with methyltransferase in vivo during productive infection, and can be methylated by HRMT1L2 (human protein arginine methyltransferase 1) in vitro. Comparative analysis of E1B-AP5 and L4-100 kDa protein methylation in Ad-infected HeLa, MCF-7 and H1299 cells revealed that the profile of protein arginine methylation correlates with the efficiency of Ad proteins production. Our results suggest that protein arginine methylation is an important host-cell function required for efficient Ad replication.
Figures




References
-
- Shenk T. Adenoviridae: the viruses and their replication. In: Fields B. N., Knipe D. M., Howley P. M., editors. Virology. New York: Lippincott-Raven; 1996. pp. 2111–2148.
-
- Flint S. J. Nature Publishing Group; 2001. Adenoviruses, Encyclopedia of Life Sciences. Electronic Citation.
-
- Dobner T., Kzhyshkowska J. Nuclear export of adenovirus RNA. Curr. Top. Microbiol. Immunol. 2001;259:25–54. - PubMed
-
- Flint S. J., Gonzalez R. A. Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 2003;272:287–330. - PubMed
-
- Kleinberger T. Induction of apoptosis by adenovirus E4orf4 protein. Apoptosis. 2000;5:211–215. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

