Rapid identification and mapping of insertion sequences in Escherichia coli genomes using vectorette PCR
- PMID: 15242519
- PMCID: PMC481064
- DOI: 10.1186/1471-2180-4-26
Rapid identification and mapping of insertion sequences in Escherichia coli genomes using vectorette PCR
Abstract
Background: Insertion sequences (IS) are small DNA segments capable of transposing within and between prokaryotic genomes, often causing insertional mutations and chromosomal rearrangements. Although several methods are available for locating ISs in microbial genomes, they are either labor-intensive or inefficient. Here, we use vectorette PCR to identify and map the genomic positions of the eight insertion sequences (IS1, 2, 3, 4, 5, 30, 150, and 186) found in E. coli strain CGSC6300, a close relative of MG1655 whose genome has been sequenced.
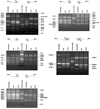
Results: Genomic DNA from strain CGSC6300 was digested with a four-base cutter Rsa I and the resulting restriction fragments ligated onto vectorette units. Using IS-specific primers directed outward from the extreme ends of each IS and a vectorette primer, flanking DNA fragments were amplified from all but one of the 37 IS elements identified in the genomic sequence of MG1655. Purification and sequencing of the PCR products confirmed that they are IS-associated flanking DNA fragments corresponding to the known IS locations in the MG1655 genome. Seven additional insertions were found in strain CGSC6300 indicating that very closely related isolates of the same laboratory strain (the K12 isolate) may differ in their IS complement. Two other E. coli K12 derivatives, TD2 and TD10, were also analyzed by vectorette PCR. They share 36 of the MG1655 IS sites as well as having 16 and 18 additional insertions, respectively.
Conclusion: This study shows that vectorette PCR is a swift, efficient, reliable method for typing microbial strains and identifying and mapping IS insertion sites present in microbial genomes. Unlike Southern hybridization and inverse PCR, our approach involves only one genomic digest and one ligation step. Vectorette PCR is then used to simultaneously amplify all IS elements of a given type, making it a rapid and sensitive means to survey IS elements in genomes. The ability to rapidly identify the IS complements of microbial genomes should facilitate subtyping closely related pathogens during disease outbreaks.
Figures


Similar articles
-
A genomic library-based amplification approach (GL-PCR) for the mapping of multiple IS6110 insertion sites and strain differentiation of Mycobacterium tuberculosis.J Microbiol Methods. 2006 Nov;67(2):202-11. doi: 10.1016/j.mimet.2006.03.021. Epub 2006 May 24. J Microbiol Methods. 2006. PMID: 16725220
-
Design and evaluation of PCR primers which differentiate Escherichia coli O157:H7 and related serotypes.J Appl Microbiol. 2009 Jan;106(1):149-60. doi: 10.1111/j.1365-2672.2008.03987.x. Epub 2008 Dec 19. J Appl Microbiol. 2009. PMID: 19120620
-
Rapid identification of Escherichia coli safety and laboratory strain lineages based on Multiplex-PCR.FEMS Microbiol Lett. 2007 Apr;269(1):36-40. doi: 10.1111/j.1574-6968.2006.00594.x. FEMS Microbiol Lett. 2007. PMID: 17343689
-
Insertion of in-frame sequence tags into proteins using transposons.Methods. 2000 Jan;20(1):55-61. doi: 10.1006/meth.1999.0905. Methods. 2000. PMID: 10610804 Review.
-
[E coli genomes].Tanpakushitsu Kakusan Koso. 1998 Aug;43(10):1381-7. Tanpakushitsu Kakusan Koso. 1998. PMID: 9867604 Review. Japanese. No abstract available.
Cited by
-
Highly sensitive real-time PCR for specific detection and quantification of Coxiella burnetii.BMC Microbiol. 2006 Jan 19;6:2. doi: 10.1186/1471-2180-6-2. BMC Microbiol. 2006. PMID: 16423303 Free PMC article.
-
InMut-finder: a software tool for insertion identification in mutagenesis using Nanopore long reads.BMC Genomics. 2021 Dec 19;22(1):908. doi: 10.1186/s12864-021-08206-9. BMC Genomics. 2021. PMID: 34923956 Free PMC article.
-
Metabolic stress constrains microbial L-cysteine production in Escherichia coli by accelerating transposition through mobile genetic elements.Microb Cell Fact. 2023 Jan 16;22(1):10. doi: 10.1186/s12934-023-02021-5. Microb Cell Fact. 2023. PMID: 36642733 Free PMC article.
-
Rates of transposition in Escherichia coli.Biol Lett. 2013 Dec 4;9(6):20130838. doi: 10.1098/rsbl.2013.0838. Print 2013. Biol Lett. 2013. PMID: 24307531 Free PMC article.
-
Genome-wide localization of mobile elements: experimental, statistical and biological considerations.BMC Genomics. 2005 Jun 1;6:81. doi: 10.1186/1471-2164-6-81. BMC Genomics. 2005. PMID: 15929794 Free PMC article.
References
-
- Blot M. Transposable elements and adaptation of host bacteria. Genetica. 1994;93:5–12. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

