Assessment of partial sequencing of the 65-kilodalton heat shock protein gene (hsp65) for routine identification of Mycobacterium species isolated from clinical sources
- PMID: 15243051
- PMCID: PMC446301
- DOI: 10.1128/JCM.42.7.3000-3011.2004
Assessment of partial sequencing of the 65-kilodalton heat shock protein gene (hsp65) for routine identification of Mycobacterium species isolated from clinical sources
Abstract
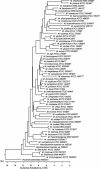
We assessed the ability of an in-house database, consisting of 111 hsp65 sequences from putative and valid Mycobacterium species or described groups, to identify 689 mycobacterial clinical isolates from 35 species or groups. A preliminary assessment indicated that hsp65 sequencing confirmed the identification of 79.4% of the isolates from the 32 species examined, including all Mycobacterium tuberculosis complex isolates, all isolates from 13 other species, and 95.6% of all M. avium-M. intracellulare complex isolates. Identification discrepancies were most frequently encountered with isolates submitted as M. chelonae, M. fortuitum, M. gordonae, M. scrofulaceum, and M. terrae. Reexamination of isolates with discrepant identifications confirmed that hsp65 identifications were correct in a further 40 isolates. This brought the overall agreement between hsp65 sequencing and the other identification methods to 85.2%. The remaining 102 isolates had sequence matches below our acceptance criterion, had nondifferential sequence matches between two or more species, were identified by 16S rRNA sequencing as a putative taxonomic group not contained in our database, or were identified by hsp65 and 16S rRNA gene sequencing as a species not in our biochemical test database or had conflicting identifications. Therefore, to incorporate the unconfirmed isolates it was necessary to create 29 additional entries in our hsp65 identification database: 18 associated with valid species, 7 indicating unique sequences not associated with valid or putative species or groups, and 4 associated with unique, but currently described taxonomic groups. Confidence in the hsp65 sequence identification of a clinical isolate is best when sequence matches of 100% occur, but our data indicate that correct identifications can be confidently made when unambiguous matches exceeding 97% occur, but are dependent on the completeness of the database. Our study indicates that for hsp65 sequencing to be an effective means for identifying mycobacteria a comprehensive database must be constructed. hsp65 sequencing has the advantage of being more rapid and less expensive than biochemical test panels, uses a single set of reagents to identify both rapid- and slow-growing mycobacteria, and can provide a more definitive identification.
Figures

References
-
- Alcaide, F., I. Richter, C. Bernasconi, B. Springer, C. Hagenau, R. Schulze-Robbecke, E. Tortoli, R. Martin, E. C. Bottger, and A. Telenti. 1997. Heterogeneity and clonality among isolates of Mycobacterium kansasii: implications for epidemiological and pathogenicity studies. J. Clin. Microbiol. 35:1959-1964. - PMC - PubMed
-
- Brown, B. A., B. Springer, V. A. Steingrube, R. W. Wilson, G. E. Pfyffer, M. J. Garcia, M. C. Menendez, B. Rodriguez-Salgado, K. C. Jost, Jr., S. H. Chiu, G. O. Onyi, E. C. Böttger, and R. J. Wallace, Jr. 1999. Mycobacterium wolinskyi sp. nov. and Mycobacterium goodii sp. nov. rapidly growing species related to Mycobacterium smegmatis and associated with human wound infections: a cooperative study from the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 49:1493-1511. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

