Quantitative analysis of diverse Lactobacillus species present in advanced dental caries
- PMID: 15243071
- PMCID: PMC446321
- DOI: 10.1128/JCM.42.7.3128-3136.2004
Quantitative analysis of diverse Lactobacillus species present in advanced dental caries
Abstract
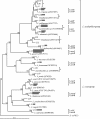
Our previous analysis of 65 advanced dental caries lesions by traditional culture techniques indicated that lactobacilli were numerous in the advancing front of the progressive lesion. Production of organic acids by lactobacilli is considered to be important in causing decalcification of the dentinal matrix. The present study was undertaken to define more precisely the diversity of lactobacilli found in this environment and to quantify the major species and phylotypes relative to total load of lactobacilli by real-time PCR. Pooled DNA was amplified by PCR with Lactobacillus genus-specific primers for subsequent cloning, sequencing, and phylogenetic analysis. Based on 16S ribosomal DNA sequence comparisons, 18 different phylotypes of lactobacilli were detected, including strong representation of both novel and gastrointestinal phylotypes. Specific PCR primers were designed for nine prominent species, including Lactobacillus gasseri, L. ultunensis, L. salivarius, L. rhamnosus, L. casei, L. crispatus, L. delbrueckii, L. fermentum, and L. gallinarum. More than three different species were identified as being present in most of the dentine samples, confirming the widespread distribution and numerical importance of various Lactobacillus spp. in carious dentine. Quantification by real-time PCR revealed various proportions of the nine species colonizing carious dentine, with higher mean loads of L. gasseri and L. ultunensis than of the other prevalent species. The findings provide a basis for further characterization of the pathogenicity of Lactobacillus spp. in the context of extension of the carious lesion.
Figures

References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Ando, N., and E. Hoshino. 1990. Predominant obligate anaerobes invading the deep layers of root canal dentin. Int. Endodont. J. 23:20-27. - PubMed
-
- Bjorndal, L., and T. Larsen. 2000. Changes in the cultivable flora in deep carious lesions following a stepwise excavation procedure. Caries Res. 34:502-508. - PubMed
-
- Botha, S. J., S. C. Boy, F. S. Botha, and R. Senekal. 1998. Lactobacillus species associated with active caries lesions. J. Dent. Assoc. S. Afr. 53:3-6. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

