Amplicon sequencing and improved detection of human rhinovirus in respiratory samples
- PMID: 15243084
- PMCID: PMC446277
- DOI: 10.1128/JCM.42.7.3212-3218.2004
Amplicon sequencing and improved detection of human rhinovirus in respiratory samples
Erratum in
- J Clin Microbiol. 2005 Jul;43(7):3593
Abstract
Improved knowledge of the genotypic characteristics of human rhinovirus (HRV) is required, as are nucleic detection assays with the capacity to overcome both the similarities between members of the family Picornaviridae and the wide diversity of different HRV serotypes. The goal of the present study was to investigate the variability and the genotypic diversity of clinical strains circulating in the community. Since most reverse transcription (RT)-PCR assays available cannot differentiate HRV from other members of the family Picornaviridae, we also validated an assay specific for HRV detection. The 5' noncoding regions of 87 different HRV serotypes and clinical isolates were sequenced. On the basis of sequence analysis and phylogenetic determination, we first confirmed that all clinical isolates were HRV. We then validated a real-time RT-PCR assay that was able not only to detect all HRV serotypes and all clinical isolates tested but also to accurately discriminate between rhinovirus and other viruses from the family Picornaviridae. This assay was negative with isolates of coxsackievirus (types A and B), echovirus, enterovirus, parechovirus, and poliovirus, as well as nonpicornaviruses. Among a series of bronchoalveolar lavage specimens, 4% (7 of 161) were positive by culture, whereas 13% (21 of 161) were positive by RT-PCR. In the present study we showed that to specifically identify HRV in clinical specimens, diagnostic assays need to overcome both the diversities and the similarities of picornaviruses. By sequencing the 5' noncoding regions of rhinoviruses recovered from clinical specimens, we designed probes that could specifically detect rhinovirus.
Figures



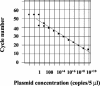
References
-
- Arruda, E., T. R. Boyle, B. Winther, D. C. Pevear, J. M. J. Gwaltney, and F. G. Hayden. 1995. Localization of human rhinovirus replication in the upper respiratory tract by in situ hybridization. J. Infect. Dis. 171:1329-1333. - PubMed
-
- Arruda, E., and F. G. Hayden. 1993. Detection of human rhinovirus RNA in nasal washings by PCR. Mol. Cell. Probes 7:373-379. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

