A GFP-based assay for rapid screening of compounds affecting ARE-dependent mRNA turnover
- PMID: 15247322
- PMCID: PMC443554
- DOI: 10.1093/nar/gnh086
A GFP-based assay for rapid screening of compounds affecting ARE-dependent mRNA turnover
Abstract
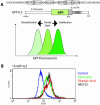
A reporter transcript containing the green fluorescent protein (GFP) gene upstream of the destabilizing 3'-untranslated region (3'-UTR) of the murine IL-3 gene was inserted in mouse PB-3c-15 mast cells. The GFP-IL-3 transcript was inherently unstable due to the presence of an adenosine-uridine (AU)-rich element (ARE) in the 3'-UTR and was subject to rapid decay giving a low baseline of GFP fluorescence. Transcript stabilization with ionomycin resulted in an increase of fluorescence that is quantitated by FACS analysis of responding cells. Using this system we have identified okadaic acid as a novel stabilizing compound, and investigated the upstream signaling pathways leading to stabilization. This reporter system has the advantage of speed and simplicity over standard methods currently in use and in addition to serving as a research tool it can be easily automated to increase throughput for drug discovery.
Figures





References
-
- Mitchell P. and Tollervey,D. (2000) mRNA stability in eukaryotes. Curr. Opin. Genet. Dev., 10, 193–198. - PubMed
-
- Guhayinogi J. and Brewer,G. (2001) Regulation of mRNA stability in mammalian cells. Gene, 265, 11–23. - PubMed
-
- Nair A.P.K., Hirsch,H.H. and Moroni,C. (1992) Mast cells sensitive to v-H-ras transformation are hyperinducible for IL-3 expression and have lost tumor-suppressor activity. Oncogene, 7, 1963–1972. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

