Packaging motor from double-stranded RNA bacteriophage phi12 acts as an obligatory passive conduit during transcription
- PMID: 15247341
- PMCID: PMC484169
- DOI: 10.1093/nar/gkh680
Packaging motor from double-stranded RNA bacteriophage phi12 acts as an obligatory passive conduit during transcription
Abstract
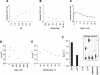
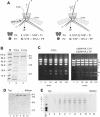
Double-stranded RNA viruses sequester their genomes within a protein shell, called the polymerase complex. Translocation of ssRNA into (packaging) and out (transcription) of the polymerase complex are essential steps in the life cycle of the dsRNA bacteriophages of the Cystoviridae family (phi6-phi14). Both processes require a viral molecular motor P4, an NTPase, which bears structural and functional similarities to hexameric helicases. In effect, switching between the packaging and the transcription mode requires the translocation direction of the P4 motor to reverse. However, the mechanism of the reversal remains elusive. Here we characterize the P4 protein from bacteriophage phi12 and exploit its purine nucleotide specificity to delineate P4 role in transcription. The results indicate that while P4 actively translocates RNA during packaging it acts as a passive conduit for RNA export. The directionality switching is accomplished via the regulation of P4 NTPase activity within the polymerase core.
Figures

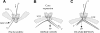
Similar articles
-
Characterization of phi12, a bacteriophage related to phi6: nucleotide sequence of the large double-stranded RNA.Virology. 2002 Apr 10;295(2):266-71. doi: 10.1006/viro.2002.1436. Virology. 2002. PMID: 12033785
-
Self-assembly of double-stranded RNA bacteriophages.Virus Res. 2004 Apr;101(1):93-100. doi: 10.1016/j.virusres.2003.12.009. Virus Res. 2004. PMID: 15010220 Review.
-
RNA packaging device of double-stranded RNA bacteriophages, possibly as simple as hexamer of P4 protein.J Biol Chem. 2003 Nov 28;278(48):48084-91. doi: 10.1074/jbc.M306928200. Epub 2003 Sep 8. J Biol Chem. 2003. PMID: 12966097
-
Structure and NTPase activity of the RNA-translocating protein (P4) of bacteriophage phi 6.J Mol Biol. 1998 Jun 5;279(2):347-59. doi: 10.1006/jmbi.1998.1772. J Mol Biol. 1998. PMID: 9642042
-
RNA-dependent RNA polymerases of dsRNA bacteriophages.Virus Res. 2004 Apr;101(1):45-55. doi: 10.1016/j.virusres.2003.12.005. Virus Res. 2004. PMID: 15010216 Review.
Cited by
-
Structural Studies of Bacteriophage Φ6 and Its Transformations during Its Life Cycle.Viruses. 2023 Dec 11;15(12):2404. doi: 10.3390/v15122404. Viruses. 2023. PMID: 38140645 Free PMC article. Review.
-
Structure and dynamics of the P7 protein from the bacteriophage phi 12.J Mol Biol. 2008 Oct 3;382(2):402-22. doi: 10.1016/j.jmb.2008.07.006. Epub 2008 Jul 11. J Mol Biol. 2008. PMID: 18647606 Free PMC article.
-
Tracking in atomic detail the functional specializations in viral RecA helicases that occur during evolution.Nucleic Acids Res. 2013 Nov;41(20):9396-410. doi: 10.1093/nar/gkt713. Epub 2013 Aug 11. Nucleic Acids Res. 2013. PMID: 23939620 Free PMC article.
-
Cryo-electron tomography reveals novel features of a viral RNA replication compartment.Elife. 2017 Jun 27;6:e25940. doi: 10.7554/eLife.25940. Elife. 2017. PMID: 28653620 Free PMC article.
-
Cystoviral RNA-directed RNA polymerases: Regulation of RNA synthesis on multiple time and length scales.Virus Res. 2017 Apr 15;234:135-152. doi: 10.1016/j.virusres.2017.01.006. Epub 2017 Jan 16. Virus Res. 2017. PMID: 28104452 Free PMC article. Review.
References
-
- Delagoutte E. and von Hippel,P.H. (2003) Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: integration of helicases into cellular processes. Quart. Rev. Biophys., 36, 1–69. - PubMed
-
- Caruthers J.M. and McKay,D.B. (2002) Helicase structure and mechanism. Curr. Opin. Struct. Biol., 12, 123–133. - PubMed
-
- Patel S.S. and Picha,K.M. (2000) Structure and function of hexameric helicases. Annu. Rev. Biochem., 69, 651–697. - PubMed
-
- Skordalakes E. and Berger,J.M. (2003) Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell, 114, 135–146. - PubMed
-
- Catalano C.E., Cue,D. and Feiss,M. (1995) Virus DNA packaging: the strategy used by phage lambda. Mol. Microbiol., 16, 1075–1086. - PubMed

