Comparative genomics in salt tolerance between Arabidopsis and aRabidopsis-related halophyte salt cress using Arabidopsis microarray
- PMID: 15247402
- PMCID: PMC519083
- DOI: 10.1104/pp.104.039909
Comparative genomics in salt tolerance between Arabidopsis and aRabidopsis-related halophyte salt cress using Arabidopsis microarray
Abstract
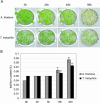
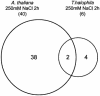
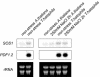
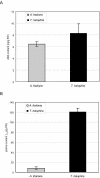
Salt cress (Thellungiella halophila), a halophyte, is a genetic model system with a small plant size, short life cycle, copious seed production, small genome size, and an efficient transformation. Its genes have a high sequence identity (90%-95% at cDNA level) to genes of its close relative, Arabidopsis. These qualities are advantageous not only in genetics but also in genomics, such as gene expression profiling using Arabidopsis cDNA microarrays. Although salt cress plants are salt tolerant and can grow in 500 mm NaCl medium, they do not have salt glands or other morphological alterations either before or after salt adaptation. This suggests that the salt tolerance in salt cress results from mechanisms that are similar to those operating in glycophytes. To elucidate the differences in the regulation of salt tolerance between salt cress and Arabidopsis, we analyzed the gene expression profiles in salt cress by using a full-length Arabidopsis cDNA microarray. In salt cress, only a few genes were induced by 250 mm NaCl stress in contrast to Arabidopsis. Notably a large number of known abiotic- and biotic-stress inducible genes, including Fe-SOD, P5CS, PDF1.2, AtNCED, P-protein, beta-glucosidase, and SOS1, were expressed in salt cress at high levels even in the absence of stress. Under normal growing conditions, salt cress accumulated Pro at much higher levels than did Arabidopsis, and this corresponded to a higher expression of AtP5CS in salt cress, a key enzyme of Pro biosynthesis. Furthermore, salt cress was more tolerant to oxidative stress than Arabidopsis. Stress tolerance of salt cress may be due to constitutive overexpression of many genes that function in stress tolerance and that are stress inducible in Arabidopsis.
Figures


Similar articles
-
Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles.Plant Physiol. 2004 Jul;135(3):1718-37. doi: 10.1104/pp.104.041723. Epub 2004 Jul 9. Plant Physiol. 2004. PMID: 15247369 Free PMC article.
-
Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T. halophila.Plant Cell Environ. 2006 Jul;29(7):1220-34. doi: 10.1111/j.1365-3040.2006.01502.x. Plant Cell Environ. 2006. PMID: 17080945
-
Structural analysis of 83-kb genomic DNA from Thellungiella halophila: sequence features and microcolinearity between salt cress and Arabidopsis thaliana.Genomics. 2009 Nov;94(5):324-32. doi: 10.1016/j.ygeno.2009.07.006. Epub 2009 Jul 29. Genomics. 2009. PMID: 19646522
-
Halophytism: What Have We Learnt From Arabidopsis thaliana Relative Model Systems?Plant Physiol. 2018 Nov;178(3):972-988. doi: 10.1104/pp.18.00863. Epub 2018 Sep 20. Plant Physiol. 2018. PMID: 30237204 Free PMC article. Review.
-
Adaptative Mechanisms of Halophytic Eutrema salsugineum Encountering Saline Environment.Front Plant Sci. 2022 Jun 28;13:909527. doi: 10.3389/fpls.2022.909527. eCollection 2022. Front Plant Sci. 2022. PMID: 35837468 Free PMC article. Review.
Cited by
-
Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency.PLoS One. 2012;7(2):e32124. doi: 10.1371/journal.pone.0032124. Epub 2012 Feb 15. PLoS One. 2012. PMID: 22355415 Free PMC article.
-
Salt Induces Features of a Dormancy-Like State in Seeds of Eutrema (Thellungiella) salsugineum, a Halophytic Relative of Arabidopsis.Front Plant Sci. 2016 Aug 3;7:1071. doi: 10.3389/fpls.2016.01071. eCollection 2016. Front Plant Sci. 2016. PMID: 27536302 Free PMC article.
-
Different transcriptional response to Xanthomonas citri subsp. citri between kumquat and sweet orange with contrasting canker tolerance.PLoS One. 2012;7(7):e41790. doi: 10.1371/journal.pone.0041790. Epub 2012 Jul 26. PLoS One. 2012. PMID: 22848606 Free PMC article.
-
Transcriptomic profiling of the salt-stress response in the wild recretohalophyte Reaumuria trigyna.BMC Genomics. 2013 Jan 16;14:29. doi: 10.1186/1471-2164-14-29. BMC Genomics. 2013. PMID: 23324106 Free PMC article.
-
DNA millichips as a low-cost platform for gene expression analysis.Plant Physiol. 2012 Jun;159(2):548-57. doi: 10.1104/pp.112.195230. Epub 2012 Apr 13. Plant Physiol. 2012. PMID: 22505730 Free PMC article.
References
-
- Abarca D, Roldan M, Martin M, Sabater B (2001) Arabidopsis thaliana ecotype Cvi shows an increased tolerance to photo-oxidative stress and contains a new chloroplastic copper/zinc superoxide dismutase isoenzyme. J Exp Bot 52: 1417–1425 - PubMed
-
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 - PubMed
-
- Chandok MR, Ytterberg AJ, van Wijk KJ, Klessig DF (2003) The pathogen-inducible nitric oxide synthase (iNOS) in plants is a variant of the P protein of the glycine decarboxylase complex. Cell 113: 469–482 - PubMed
-
- Deuschle K, Funck D, Hellmann H, Daschner K, Binder S, Frommer WB (2001) A nuclear gene encoding mitochondrial delta-pyrroline-5-carboxylate dehydrogenase and its potential role in protection from proline toxicity. Plant J 27: 345–356 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

