Alzheimer's brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication
- PMID: 15247418
- PMCID: PMC490002
- DOI: 10.1073/pnas.0403649101
Alzheimer's brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication
Abstract
Defects in mitochondrial oxidative phosphorylation have frequently been associated with Alzheimer's disease (AD), and both inherited and somatic mtDNA mutations have been reported in certain AD cases. To determine whether mtDNA mutations contribute more generally to the etiology of AD, we have investigated the sequence of the mtDNA control region (CR) from AD brains for possible disease-causing mutations. Sixty-five percent of the AD brains harbored the T414G mutation, whereas this mutation was absent from all controls. Moreover, cloning and sequencing of the mtDNA CR from patient and control brains revealed that all AD brains had an average 63% increase in heteroplasmic mtDNA CR mutations and that AD brains from patients 80 years and older had a 130% increase in heteroplasmic CR mutations. In addition, these mutations preferentially altered known mtDNA regulatory elements. Certain AD brains harbored the disease-specific CR mutations T414C and T477C, and several AD brains between 74 and 83 years of age harbored the CR mutations T477C, T146C, and T195C, at levels up to 70-80% heteroplasmy. AD patient brains also had an average 50% reduction in the mtDNA L-strand ND6 transcript and in the mtDNA/nuclear DNA ratio. Because reduced ND6 mRNA and mtDNA copy numbers would reduce brain oxidative phosphorylation, these CR mutations could account for some of the mitochondrial defects observed in AD.
Figures
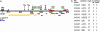

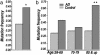

References
-
- Tandon, A., Rogaeva, E., Mullan, M. & St. George-Hyslop, P. H. (2000) Curr. Opin. Neurol. 13, 377-384. - PubMed
-
- Smith, M. A., Casadesus, G., Joseph, J. A. & Perry, G. (2002) Free Radical Biol. Med. 33, 1194-1199. - PubMed
-
- Castellani, R., Hirai, K., Aliev, G., Drew, K. L., Nunomura, A., Takeda, A., Cash, A. D., Obrenovich, M. E., Perry, G. & Smith, M. A. (2002) J. Neurosci. Res. 70, 357-360. - PubMed
-
- Harman, D. (2002) Ann. N.Y. Acad. Sci. 959, 384-395. - PubMed
-
- Cottrell, D. A., Borthwick, G. M., Johnson, M. A., Ince, P. G. & Turnbull, D. M. (2002) Neuropathol. Appl. Neurobiol. 28, 390-396. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

