Origin, evolution, and mechanism of 5' tRNA editing in chytridiomycete fungi
- PMID: 15247432
- PMCID: PMC1370609
- DOI: 10.1261/rna.7330504
Origin, evolution, and mechanism of 5' tRNA editing in chytridiomycete fungi
Abstract
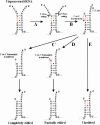
5' tRNA editing has been demonstrated to occur in the mitochondria of the distantly related rhizopod amoeba Acanthamoeba castellanii and the chytridiomycete fungus Spizellomyces punctatus. In these organisms, canonical tRNA structures are restored by removing mismatched nucleotides at the first three 5' positions and replacing them with nucleotides capable of forming Watson-Crick base pairs with their 3' counterparts. This form of editing seems likely to occur in members of Amoebozoa other than A. castellanii, as well as in members of Heterolobosea. Evidence for 5' tRNA editing has not been found to date, however, in any other fungus including the deeply branching chytridiomycete Allomyces macrogynus. We predicted that a similar form of tRNA editing would occur in members of the chytridiomycete order Monoblepharidales based on the analysis of complete mitochondrial tRNA complements. This prediction was confirmed by analysis of tRNA sequences using a tRNA circularization/RT-PCR-based approach. The presence of partially and completely unedited tRNAs in members of the Monoblepharidales suggests the involvement of a 5'-to-3' exonuclease rather than an endonuclease in removing the three 5' nucleotides from a tRNA substrate. Surprisingly, analysis of the mtDNA of the chytridiomycete Rhizophydium brooksianum, which branches as a sister group to S. punctatus in molecular phylogenies, did not suggest the presence of editing. This prediction was also confirmed experimentally. The absence of tRNA editing in R. brooksianum raises the possibility that 5' tRNA editing may have evolved twice independently within Chytridiomycota, once in the lineage leading to S. punctatus and once in the lineage leading to the Monoblepharidales.
Figures


References
-
- Baldauf, S.L., Roger, A.J., Wenk-Siefert, I., and Doolittle, W.F. 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290: 972–977. - PubMed
-
- Barth, C., Greferath, U., Kotsifas, M., and Fisher, P.R. 1999. Polycistronic transcription and editing of the mitochondrial small subunit (SSU) ribosomal RNA in Dictyostelium discoideum. Curr. Genet. 36: 55–61. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
