Mutation in mitochondrial complex I ND6 subunit is associated with defective response to hypoxia in human glioma cells
- PMID: 15248896
- PMCID: PMC481082
- DOI: 10.1186/1476-4598-3-19
Mutation in mitochondrial complex I ND6 subunit is associated with defective response to hypoxia in human glioma cells
Abstract
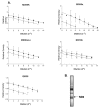
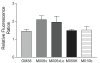
Background: Hypoxia-tolerant human glioma cells reduce oxygen consumption rate in response to oxygen deficit, a defense mechanism that contributes to survival under moderately hypoxic conditions. In contrast, hypoxia-sensitive cells lack this ability. As it has been previously shown that hypoxia-tolerant (M006x, M006xLo, M059K) and -sensitive (M010b) glioma cells express differences in mitochondrial function, we investigated whether mitochondrial DNA-encoded mutations are associated with differences in the initial response to oxygen deficit.
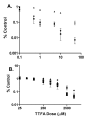
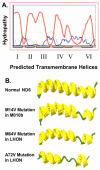
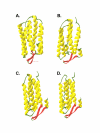
Results: The mitochondrial genome was sequenced and 23 mtDNA alterations were identified, one of which was an unreported mutation (T-C transition in base pair 14634) in the hypoxia-sensitive cell line, M010b, that resulted in a single amino acid change in the gene encoding the ND6 subunit of NADH:ubiquinone oxidoreductase (Complex I). The T14634C mutation did not abrogate ND6 protein expression, however, M010b cells were more resistant to rotenone, an agent used to screen for Complex I mutations, and adriamycin, an agent activated by redox cycling. The specific function of mtDNA-encoded, membrane-embedded Complex I ND subunits is not known at present. Current models suggest that the transmembrane arm of Complex I may serve as a conformationally driven proton channel. As cellular respiration is regulated, in part, by proton flux, we used homology-based modeling and computational molecular biology to predict the 3D structure of the wild type and mutated ND6 proteins. These models predict that the T14634C mutation alters the structure and orientation of the trans-membrane helices of the ND6 protein.
Conclusion: Complex I ND subunits are mutational hot spots in tumor mtDNA. Genetic changes that alter Complex I structure and function may alter a cell's ability to respond to oxygen deficit and consolidate hypoxia rescue mechanisms, and may contribute to resistance to chemotherapeutic agents that require redox cycling for activation.
Figures


References
-
- Dachs GU, Chaplin DJ. Microenvironmental control of gene expression: implications for tumor angiogenesis, progression, and metastasis. Semin Radiat Oncol. 1998;8:208–216. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

