Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications
- PMID: 15249599
- PMCID: PMC484200
- DOI: 10.1093/nar/gnh101
Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications
Abstract
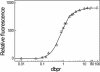
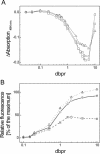
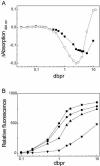
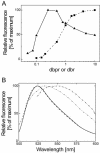
The detection of double-stranded (ds) DNA by SYBR Green I (SG) is important in many molecular biology methods including gel electrophoresis, dsDNA quantification in solution and real-time PCR. Biophysical studies at defined dye/base pair ratios (dbprs) were used to determine the structure-property relationships that affect methods applying SG. These studies revealed the occurrence of intercalation, followed by surface binding at dbprs above approximately 0.15. Only the latter led to a significant increase in fluorescence. Studies with poly(dA)* poly(dT) and poly(dG)* poly(dC) homopolymers showed sequence-specific binding of SG. Also, salts had a marked impact on SG fluorescence. We also noted binding of SG to single-stranded (ss) DNA, although SG/ssDNA fluorescence was at least approximately 11-fold lower than with dsDNA. To perform these studies, we determined the structure of SG by mass spectrometry and NMR analysis to be [2-[N-(3-dimethylaminopropyl)-N-propylamino]-4-[2,3-dihydro-3-methyl-(benzo-1,3-thiazol-2-yl)-methylidene]-1-phenyl-quinolinium]. For comparison, the structure of PicoGreen (PG) was also determined and is [2-[N-bis-(3-dimethylaminopropyl)-amino]-4-[2,3-dihydro-3-methyl-(benzo-1,3-thiazol-2-yl)-methylidene]-1-phenyl-quinolinium]+. These structure-property relationships help in the design of methods that use SG, in particular dsDNA quantification in solution and real-time PCR.
Figures
Similar articles
-
Bisintercalation of homodimeric thiazole orange dyes in DNA: effect of modifying the linker.Bioconjug Chem. 1997 Nov-Dec;8(6):869-77. doi: 10.1021/bc970067k. Bioconjug Chem. 1997. PMID: 9404660
-
Characterization of PicoGreen reagent and development of a fluorescence-based solution assay for double-stranded DNA quantitation.Anal Biochem. 1997 Jul 1;249(2):228-38. doi: 10.1006/abio.1997.2177. Anal Biochem. 1997. PMID: 9212875
-
Labeling-free fluorescent detection of DNA hybridization through FRET from pyrene excimer to DNA intercalator SYBR green I.Biosens Bioelectron. 2015 Mar 15;65:103-7. doi: 10.1016/j.bios.2014.10.029. Epub 2014 Oct 18. Biosens Bioelectron. 2015. PMID: 25461145
-
Drug-DNA sequence-dependent interactions analysed by electric linear dichroism.J Mol Recognit. 1992 Dec;5(4):155-71. doi: 10.1002/jmr.300050406. J Mol Recognit. 1992. PMID: 1339484 Review.
-
PicoGreen assay for nucleic acid quantification - Applications, challenges, and solutions.Anal Biochem. 2024 Sep;692:115577. doi: 10.1016/j.ab.2024.115577. Epub 2024 May 22. Anal Biochem. 2024. PMID: 38789006 Review.
Cited by
-
PicoGreen assay of circular DNA for radiation biodosimetry.Radiat Res. 2015 Feb;183(2):188-95. doi: 10.1667/RR13556.1. Epub 2015 Jan 9. Radiat Res. 2015. PMID: 25574588 Free PMC article.
-
Fluorescent properties of pentamethine cyanine dyes with cyclopentene and cyclohexene group in presence of biological molecules.J Fluoresc. 2005 Nov;15(6):849-57. doi: 10.1007/s10895-005-0002-7. J Fluoresc. 2005. PMID: 16283530
-
Fluorescence-based Broad Dynamic Range Viscosity Probes.J Fluoresc. 2014 Mar;24(2):397-402. doi: 10.1007/s10895-013-1304-9. Epub 2013 Nov 16. J Fluoresc. 2014. PMID: 24241893
-
PCR inhibition in qPCR, dPCR and MPS-mechanisms and solutions.Anal Bioanal Chem. 2020 Apr;412(9):2009-2023. doi: 10.1007/s00216-020-02490-2. Epub 2020 Feb 12. Anal Bioanal Chem. 2020. PMID: 32052066 Free PMC article. Review.
-
Fluorescence characterization of the structural heterogeneity of polytene chromosomes.J Fluoresc. 2010 Jan;20(1):37-41. doi: 10.1007/s10895-009-0519-2. Epub 2009 Jul 24. J Fluoresc. 2010. PMID: 19629653
References
-
- Vitzthum F. and Bernhagen,J. (2002) SYBR Green I: an ultrasensitive fluorescent dye for double-standed DNA quantification in solution and other applications. Recent Res. Devel. Anal. Biochem., 2, 65–93.
-
- Haugland R.P. (2001) Handbook of Fluorescent Probes and Research Chemicals, 8th edn. Molecular Probes, Inc., Eugene, OR.
-
- Schneeberger C., Speiser,P., Kury,F. and Zeillinger,R. (1995) Quantitative detection of reverse transcriptase–PCR products by means of a novel and sensitive DNA stain. PCR Methods Appl., 4, 234–238. - PubMed
-
- Jin X., Dong,F. and Singer,V.L. (1996) SYBR Green I nucleic acid gel stain provides a sensitive fluorescent method for detecting gel mobility shift products. FASEB J., 10, A1128.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

