Pattern of polymorphism after strong artificial selection in a domestication event
- PMID: 15249682
- PMCID: PMC489992
- DOI: 10.1073/pnas.0401720101
Pattern of polymorphism after strong artificial selection in a domestication event
Abstract
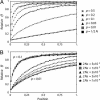
The process of strong artificial selection during a domestication event is modeled, and its effect on the pattern of DNA polymorphism is investigated. The model also considers population bottleneck during domestication. Artificial selection during domestication is different from a regular selective sweep because artificial selection acts on alleles that may have been neutral variants before domestication. Therefore, the fixation of such a beneficial allele does not always wipe out DNA variation in the surrounding region. The amount by which variation is reduced largely depends on the initial frequency of the beneficial allele, p. As a consequence, p has a strong effect on the likelihood of detecting the signature of selection during domestication from patterns of polymorphism. These theoretical results are discussed in light of data collected from maize. Although the main focus of this article is on domestication, this model can also be generalized to describe selective sweeps from standing genetic variation.
Figures




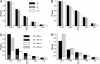
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

