Two-fold differences are the detection limit for determining transgene copy numbers in plants by real-time PCR
- PMID: 15251044
- PMCID: PMC493272
- DOI: 10.1186/1472-6750-4-14
Two-fold differences are the detection limit for determining transgene copy numbers in plants by real-time PCR
Abstract
Background: After transformation, plants that are homozygous and contain one copy of the transgene are typically selected for further study. If real-time PCR is to be used to determine copy number and zygosity, it must be able to distinguish hemizygous from homozygous and one-copy from two-copy plants. That is, it must be able to detect two-fold differences.
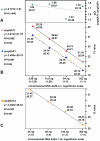
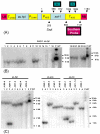
Results: When transgenic Nicotiana attenuata plants which had been previously determined by Southern analysis to contain one or two copies of the transgene, were analyzed by real-time PCR (2-delta delta Ct method), the method failed to confirm the results from the Southern analysis. In a second data set we analyzed offspring of a hemizygous one-copy plant, which were expected to segregate into three groups of offspring in a 1:2:1 ratio: no transgene, hemizygous, homozygous. Because it was not possible to distinguish homozygous from hemizygous plants with real-time PCR, we could not verify this segregation ratio.
Conclusions: Detection of two-fold differences by real-time PCR is essential if this procedure is to be used for the characterization of transgenic plants. However, given the high variability between replicates, a detection of two-fold differences is in many cases not possible; in such cases Southern analysis is the more reliable procedure.
Figures
References
-
- Holck A, Va M, Didierjean L, Rudi K. 5 '-nuclease PCR for quantitative event-specific detection of the genetically modified Mon810 MaisGard maize. Eur Food Res Technol. 2002;214:449–453. doi: 10.1007/s00217-001-0473-y. - DOI
-
- Ingham DJ, Beer S, Money S, Hansen G. Quantitative real-time PCR assay for determining transgene copy number in transformed plants. Biotechniques. 2001;31:132–140. - PubMed
-
- Song P, Cai CQ, Skokut M, Kosegi BD, Petolino JF. Quantitative real-time PCR as a screening tool for estimating transgene copy number in WHISKERS (TM)-derived transgenic maize. Plant Cell Reports. 2002;20:948–954. doi: 10.1007/s00299-001-0432-x. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

