ChIP Display: novel method for identification of genomic targets of transcription factors
- PMID: 15252151
- PMCID: PMC484196
- DOI: 10.1093/nar/gnh097
ChIP Display: novel method for identification of genomic targets of transcription factors
Abstract
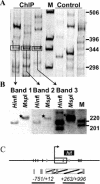
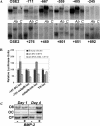
Novel protein-DNA interactions in mammalian cells are traditionally discovered in the course of promoter studies. The genomic era presents opportunities for the reverse; namely, the discovery of novel target genes for transcription factors of interest. Chromatin immunoprecipitation (ChIP) is typically used to test whether a protein binds to a candidate promoter in living cells. We developed a new method, ChIP Display (CD), which allows genome-wide unbiased identification of target genes occupied by transcription factors of interest. Initial CD experiments pursuing target genes for RUNX2, an osteoblast master transcription factor, have already resulted in the identification of four genes that had never been reported as targets of RUNX2. One of them, Osbpl8, was subjected to mRNA and promoter-reporter analyses, which provided functional proof for its regulation by RUNX2. CD will help to assemble the puzzle of interactions between transcription factors and the genome.
Figures



Similar articles
-
Identification of transcription factor target genes by ChIP display.Methods Mol Biol. 2008;455:177-90. doi: 10.1007/978-1-59745-104-8_14. Methods Mol Biol. 2008. PMID: 18463820
-
Identification of novel protein/DNA interactions within the promoter of the bone-related transcription factor Runx2/Cbfa1.J Cell Biochem. 2002;86(2):403-12. doi: 10.1002/jcb.10238. J Cell Biochem. 2002. PMID: 12112009
-
Computer-assisted identification of cell cycle-related genes: new targets for E2F transcription factors.J Mol Biol. 2001 May 25;309(1):99-120. doi: 10.1006/jmbi.2001.4650. J Mol Biol. 2001. PMID: 11491305
-
Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors.Crit Rev Eukaryot Gene Expr. 2004;14(1-2):1-41. Crit Rev Eukaryot Gene Expr. 2004. PMID: 15104525 Review.
-
Use of chromatin immunoprecipitation assays in genome-wide location analysis of mammalian transcription factors.Methods Enzymol. 2004;376:304-15. doi: 10.1016/S0076-6879(03)76020-0. Methods Enzymol. 2004. PMID: 14975314 Review. No abstract available.
Cited by
-
Identification of novel androgen receptor target genes in prostate cancer.Mol Cancer. 2007 Jun 6;6:39. doi: 10.1186/1476-4598-6-39. Mol Cancer. 2007. PMID: 17553165 Free PMC article.
-
Huntingtin interacting protein 1 modulates the transcriptional activity of nuclear hormone receptors.J Cell Biol. 2005 Jul 18;170(2):191-200. doi: 10.1083/jcb.200503106. J Cell Biol. 2005. PMID: 16027218 Free PMC article.
-
The current state of chromatin immunoprecipitation.Mol Biotechnol. 2010 May;45(1):87-100. doi: 10.1007/s12033-009-9239-8. Mol Biotechnol. 2010. PMID: 20077036 Review.
-
Repression of Runx2 by androgen receptor (AR) in osteoblasts and prostate cancer cells: AR binds Runx2 and abrogates its recruitment to DNA.Mol Endocrinol. 2009 Aug;23(8):1203-14. doi: 10.1210/me.2008-0470. Epub 2009 Apr 23. Mol Endocrinol. 2009. PMID: 19389811 Free PMC article.
-
Distinct profiles of REST interactions with its target genes at different stages of neuronal development.Mol Biol Cell. 2005 Dec;16(12):5630-8. doi: 10.1091/mbc.e05-07-0687. Epub 2005 Sep 29. Mol Biol Cell. 2005. PMID: 16195345 Free PMC article.
References
-
- Liang P. and Pardee,A.B. (1992) Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science, 257, 967–971. - PubMed
-
- Weinmann A.S. and Farnham,P.J. (2002) Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods, 26, 37–47. - PubMed
-
- Cawley S., Bekiranov,S., Ng,H.H., Kapranov,P., Sekinger,E.A., Kampa,D., Piccolboni,A., Sementchenko,V., Cheng,J., Williams,A.J. et al. (2004) Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell, 116, 499–509. - PubMed
-
- Komori T., Yagi,H., Nomura,S., Yamaguchi,A., Sasaki,K., Deguchi,K., Shimizu,Y., Bronson,R.T., Gao,Y.H., Inada,M. et al. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell, 89, 755–764. - PubMed

