Genome-wide operon prediction in Staphylococcus aureus
- PMID: 15252153
- PMCID: PMC484181
- DOI: 10.1093/nar/gkh694
Genome-wide operon prediction in Staphylococcus aureus
Abstract
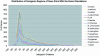
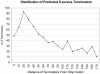
Identification of operon structure is critical to understanding gene regulation and function, and pathogenesis, and for identifying targets towards the development of new antibiotics in bacteria. Recently, the complete genome sequences of a large number of important human bacterial pathogens have become available for computational analysis, including the major human Gram-positive pathogen Staphylococcus aureus. By annotating the predicted operon structure of the S.aureus genome, we hope to facilitate the exploration of the unique biology of this organism as well as the comparative genomics across a broad range of bacteria. We have integrated several operon prediction methods and developed a consensus approach to score the likelihood of each adjacent gene pair to be co-transcribed. Gene pairs were separated into distinct operons when scores were equal to or below an empirical threshold. Using this approach, we have generated a S.aureus genome map with scores annotated at the intersections of every adjacent gene pair. This approach predicted about 864 monocistronic transcripts and 533 polycistronic operons from the protein-encoding genes in the S.aureus strain Mu50 genome. When compared with a set of experimentally determined S.aureus operons from literature sources, this method successfully predicted at least 91% of gene pairs. At the transcription unit level, this approach correctly identified at least 92% of complete operons in this dataset. This consensus approach has enabled us to predict operons with high accuracy from a genome where limited experimental evidence for operon structure is available.
Figures


References
-
- Jacob F. and Monod,J. (1961) Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol., 3, 318–356. - PubMed
-
- Burton Z.F., Gross,C.A., Watanabe,K. and Burgess,R.R. (1983) The operon that encodes the sigma subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E.coli K12. Cell, 32, 335–349. - PubMed
-
- Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

