From the Cover: The dynamics of genomic-length DNA molecules in 100-nm channels
- PMID: 15252203
- PMCID: PMC503729
- DOI: 10.1073/pnas.0403849101
From the Cover: The dynamics of genomic-length DNA molecules in 100-nm channels
Abstract
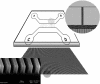
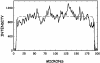
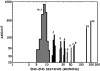
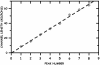
We show that genomic-length DNA molecules imaged in nanochannels have an extension along the channel that scales linearly with the contour length of the polymer, in agreement with the scaling arguments developed by de Gennes for self-avoiding confined polymers. This fundamental relationship allows us to measure directly the contour length of single DNA molecules confined in the channels, and the statistical analysis of the dynamics of the polymer in the nanochannel allows us to compute the SD of the mean of the extension. This statistical analysis allows us to measure the extension of lambda DNA multimers with a 130-nm SD in 1 min.
Figures




References
-
- Slater, G. W., Desrulsseaux, C., Hubert, S. J., Mercier, J. F., Labrie, J., Boileau, J., Tessier, F. & Pepin, M. P. (2000) Electrophoresis 21, 3873–3887. - PubMed
-
- Lin, J., Qi, R., Aston, C., Jing, J., Anantharaman, T. S., Mishra, B., White, O., Daly, M. J., Minton, K. W., Venter, J. C. & Schwartz, D. C. (1999) Science 285, 1558–1562. - PubMed
-
- Flory, P. J. (1953) Principles of Polymer Chemistry (Cornell Univ. Press, Ithaca, NY).
-
- Schaefer, D. W., Joanny, J. F. & Pincus, P. (1980) Macromolecules 13, 1280–1289.
-
- Smith, D. E., Perkins, T. T. & Chu, S. (1996) Macromolecules 29, 1372–1373.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

